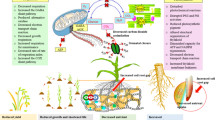
Abstract
Genetic engineering of plants for abiotic stress tolerance is a challenging task because of its multifarious nature. Comprehensive studies for develo** abiotic stress tolerance are in progress, involving genes from different pathways including osmolyte synthesis, ion homeostasis, antioxidative pathways, and regulatory genes. In the last decade, several attempts have been made to substantiate the role of “single-function” gene(s) as well as transcription factor(s) for abiotic stress tolerance. Since, the abiotic stress tolerance is multigenic in nature, therefore, the recent trend is shifting towards genetic transformation of multiple genes or transcription factors. A large number of crop plants are being engineered by abiotic stress tolerant genes and have shown the stress tolerance mostly at laboratory level. This review presents a mechanistic view of different pathways and emphasizes the function of different genes in conferring salt tolerance by genetic engineering approach. It also highlights the details of successes achieved in develo** salt tolerance in plants thus far.


Similar content being viewed by others
Abbreviations
- CBL:
-
Calcineurin B-like protein
- CIPK:
-
CBL-interacting protein kinases
- NPK1:
-
Mitogen-activated protein kinase kinase kinase
- NDPK2:
-
Nucleoside diphosphate kinase 2
- SAPK4:
-
Sucrose nonfermenting 1-related protein kinase2 (SnRK2)
- AtMEK1:
-
MAPK kinase
- MYB:
-
Myeloblastoma
- NAC:
-
No apical meristem, ATAF 1,2 and cup-shaped cotyledon
- DRE:
-
Drought responsive element
- DBF:
-
DRE binding factor
- DREB:
-
Drought responsive element binding protein
- TPS:
-
Trehalose-6-phosphate synthase
- p5cs:
-
Δ1-Pyroline-5-carboxylate synthase
- codA:
-
Choline oxidase
- BADH:
-
Betaine aldehyde dehydrogenase
- mt1D:
-
Mannitol-1-phosphate dehydrogenase
- P5CR:
-
P5C reductase
- GutD:
-
Glucitol-6-phosphate dehydrogenase
- MIPS:
-
l-Myo-Inositol-1-phosphate synthase
- APX:
-
Cytosolic ascorbate peroxidase
- DHAR:
-
Dehydroascorbate reductase
- MDHAR:
-
Mono DHAR
- SOD:
-
Superoxide dismutase
- ADC:
-
Arginine decarboxylase
- ODC:
-
Ornithine decarboxylase
- SAMDC:
-
S-Adenosyl methionine decaroboxylase
- SPDS:
-
Spermidine Synthase
- NHX-1:
-
Vacuolar Na+/H+ antiporter
- SOS1:
-
Salt overly sensitive
- TsVP:
-
H+-pyrophosphatase
- HKT2:
-
High efficiency potassium transport
References
Vinocur, B., & Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress, achievements and limitations. Current Opinion in Biotechnology, 16, 123–132.
Szabolcs, I. (1994). Salt affected soils as an ecosystems for halophytes. In V. R. Squires & A. T. Malcom (Eds.), Halophytes as a resource for livestock and for rehabilitation of degraded lands (pp. 19–24). Dordrecht: Kluwer Academic Publishers.
Niu, X., Bressan, R. A., Hasegawa, P. M., & Pardo, J. M. (1995). Ion homeostasis in NaCl stress environments. Plant Physiology, 109, 735–742.
Tester, M., & Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Annals of Botany, 91, 503–527.
Rajendran, K. A., Tester, M., & Roy, S. J. (2009). Quantifying the three main components of salinity tolerance in cereals. Plant, Cell and Environment, 32, 237–249.
Agarwal, P. K., & Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biologia Plantarum, 54, 201–212.
Shinozaki, K., & Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany, 58, 221–227.
Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2001). Transgenic approach toward develo** abiotic stress tolerance in plants. Proceedings of the Indian National Science Academy Part B, 67, 265–284.
Bhatnagar, P., Vadez, V., & Sharma, K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants, retrospect and prospects. Plant Cell Reports, 7, 411–424.
Ashraf, M., & Akram, N. A. (2009). Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnology Advances, 27, 744–752.
Shao, H. B., Chu, L. Y., Lu, Z. H., & Kang, C. M. (2008). Primary antioxidant free radical scavenging and redox signalling pathways in higher plant cells. International Journal of Biological Sciences, 4, 8–14.
Apse, M. P., & Blumwald, E. (2007). Na+ transport in plants. FEBS Letters, 581, 2247–2254.
Gaxiola, R. A., Palmgren, M. G., & Schumacher, K. (2007). Plant proton pumps. FEBS Letters, 581, 2204–2214.
Rodríguez-Rosales, M. P., Gálvez, F. J., Huertas, R., Aranda, M. N., Baghour, M., Cagnac, O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signaling and Behavior, 4, 265–276.
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology, 6, 441–445.
Agarwal, P. K., Agarwal, P., Reddy, M. K., & Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports, 25, 1263–1274.
Century, K., Reuber, T. L., & Ratcliffe, O. J. (2008). Regulating the regulators, the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiology, 147, 20–29.
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., & Shinozaki, K. (2006). Engineering drought tolerance in plants, discovering and tailoring genes unlock the future. Current Opinion in Biotechnology, 17, 113–122.
Knight, M. R., Campbell, A. K., Smith, S. M., & Trewavas, A. J. (1991). Transgenic aequorin reports the effects of touch and cold shock and elicitors on cytosolic calcium. Nature, 352, 524–526.
Knight, H., Brandt, S., & Knight, M. R. (1998). A history of stress alters drought calcium signaling pathways in Arabidopsis. The Plant Journal, 16, 681–687.
Reddy, V. S., & Reddy, A. S. (2004). Proteomics of calcium-signaling components in plants. Phytochemistry, 65, 1745–1776.
Pardo, J. M., Reddy, M. P., Yang, S., Maggio, A., Huh, G. H., Matsumoto, T., et al. (1998). Stress signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proceedings of the National Academy of Sciences of the United States of America, 96, 4718–4723. 54.
Liu, J., & Zhu, K. (1998). A calcium sensor homolog required for plant salt tolerance. Science, 280, 1943–1945.
Wang, M., Gu, D., Liu, T., Wang, Z., Guo, X., Hou, W., et al. (2007). Overexpression of a putative maize calcineurin B-like protein in Arabidopsis confers salt tolerance. Plant Molecular Biology, 65, 733–746.
Tripathi, V., Parasuraman, B., Laxmi, A., & Chattopadhyay, D. (2009). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. The Plant Journal, 58, 778–790.
Moon, H., Lee, B., Choi, G., Shin, D., Prasad, D. T., Lee, O., et al. (2003). NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences of the United States of America, 100, 358–363.
Diédhiou, C. J., Popova, O. V., Dietz, K. J., & Golldack, D. (2008). The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biology, 8, 49.
Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Molecular Cell, 15, 141–152.
Ichimura, K. (2002). Mitogen-activated protein kinase cascades in plants, a new nomenclature. Trends in Plant Science, 7, 301–308.
**ong, L., & Yang, Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid‐inducible mitogen‐activated protein kinase. The Plant Cell, 5, 745–759.
Jeong, M. J., Lee, S. K., Kim, B. G., Kwon, T. R., Cho, W. S., Park, Y. T., et al. (2006). A rice (Oryza sativa L.) MAP kinase gene, OsMAPK44, is involved in response to abiotic stresses. Plant Cell, Tissue and Organ Culture, 85, 151–160.
Kong, X., Pan, J., Zhang, M., **ng, X., Zhou, Y., Liu, Y., et al. (2011). ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant, Cell and Environment, 34, 1291–1303.
Wurzinger, B., Mair, A., Pfister, B., & Teige, M. (2011). Cross-talk of calcium dependent protein kinase and MAP kinase signaling. Plant Signaling and Behavior, 6, 8–12.
Capiati, D. A., Pais, S. M., & Tellez-Inon, M. T. (2006). Wounding increases salt tolerance in tomato plants: Evidence on the participation of calmodulin-like activities in crosstolerance signalling. Journal of Experimental Botany, 57, 2391–2400.
Reichmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors, genome-wide comparative analysis among eukaryotes. Science, 290, 2105–2110.
Shiu, S. H., Shih, M. C., & Li, W. H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiology, 139, 18–26.
Shinozaki, K., & Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature, differences and cross talk between two stress signaling pathways. Current Opinion in Plant Biology, 3, 217–223.
Grotewold, E. (2008). Transcription factors for predictive plant metabolic engineering: Are we there yet? Current Opinion in Biotechnology, 19, 138–144.
Lata, C. & Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. Journal of Experimental Botany. doi:10.1093/jxb/err210.
Gupta, K., Agarwal, P. K., Reddy, M. K., & Jha, B. (2010). SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell Reports, 29, 1131–1137.
Olsen, A. N., Ernst, H. A., Leggio, L. L., & Skriver, K. (2005). NAC transcription factors, structurally distinct, functionally diverse. Trends in Plant Science, 10, 79–87.
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., & Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis, an analysis of the cup-shaped cotyledon mutant. The Plant Cell, 9, 841–857.
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., & Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell, 85, 159–170.
He, C., Yan, J., Shen, G., Fu, L., Scott, H. A., Auld, D., et al. (2005). Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant and Cell Physiology, 46, 1848–1854.
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. The Plant Journal, 39, 863–876.
Tran, L. S., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. The Plant Journal, 49, 46–63.
Nakashima, K., Tran, L. S. P., Nguyen, D. V., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal, 51, 617–630.
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 Promoter. The Plant Cell, 16, 2481–2498.
Hu, H., Dai, M., Yao, J., **ao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America, 103, 12987–12992.
Hu, H., You, J., Fang, Y., Zhu, X., Qi, Z., & **ong, L. (2008). Characterization of a transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology, 67, 169–181.
Yokotani, N., Ichikawa, T., Kondou, Y., Matsui, M., Hirochika, H., Iwabuchi, M., et al. (2009). Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta, 229, 1065–1075.
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q. Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. The Plant Journal. 68, 302–313.
Liu, Q. L., Xu, K. D., Zhao, L. J., Pan, Y. Z., Jiang, B. B., Zhang, H. Q., et al. (2011). Overexpression of a novel chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco. Biotechnology Letters. 33, 2073–2082.
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., & Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta, 234, 331–345.
Takahashi, R., Liu, S., & Takano, T. (2009). Isolation and characterization of plasma membrane Na+/H+ antiporter genes from salt-sensitive and salt-tolerant reed plants. Journal of Plant Physiology, 166, 301–309.
Liu, X., Hong, L., Li, X. Y., Yao, Y., Hu, B., & Li, L. (2011). Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Bioscience, Biotechnology, and Biochemistry, 3, 443–450.
Yanhui, C., **aoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60, 107–124.
Dai, X. Y., Xu, Y. Y., Ma, Q. B., Xu, W. Y., Wang, T., Xue, Y. B., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology, 143, 1739–1751.
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiology, 146, 623–635.
Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B., Zhang, J. S., et al. (2008). Soybean GmMYB76, GmMYB92 and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Research, 18, 1047–1060.
Gao, J. J., Zhang, Z., Peng, R. H., **ong, A. S., Xu, J., Zhu, B., et al. (2010). Forced expression of Mdmyb10, a myb transcription factor gene from apple, enhances tolerance to osmotic stress in transgenic Arabidopsis. Molecular Biology Reports, 38, 205–211.
Mao, X., Jia, D., Li, A., Zhang, H., Tian, S., Zhang, X., et al. (2011). Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Functional & Integrative Genomics. 11, 445–465.
Liu, H., Zhou X., Dong, N., Liu, X., Zhang, H., Zhang, Z. (2011). Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Functional & Integrative Genomics. 11, 431–443.
AbuQamar, S., Luo, H., Laluk, K., Mickelbart, M. V., & Mengiste, T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. The Plant Journal, 58, 347–360.
Iturriaga, G., Suarez, R., & Nova-Franco, B. (2009). Trehalose metabolism, from osmoprotection to signaling. International Journal of Molecular Sciences, 10, 3793–3810.
Holmström, K. O., Welin, B., Mandal, A., Kristiansdottir, I., Teeri, T. H., Lamark, T., et al. (1996). Production of the Escherichia coli betaine-aldehyde dehydrogenase, an enzyme required for synthesis of the osmoprotectant glycine betaine in transgenic plants. The Plant Journal, 5, 749–758.
Pilon-Smits, E. A. H., Terry, N., Sears, T., Kim, H., Zayed, A., Hwang, S., et al. (1998). Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. Journal of Plant Physiology, 152, 525–532.
Miranda, J. A., Avonce, N., Suárez, R., Thevelein, J. M., van Dijck, P., & Iturriaga, G. (2007). A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta, 226, 1411–1421.
Suarez, R., Calderón, C., & Iturriaga, G. (2009). Improved tolerance to multiple abiotic stresses in transgenic alfalfa accumulating trehalose. Crop Science, 49, 1791–1799.
Cortina, C., & Culianez-Macia, F. A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Science, 169, 75–82.
Garg, A. K., Kim, J. K., Owens, T. G., Ranwala, A. P., Choi, Y. D., Kochian, L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proceedings of the National Academy of Sciences of the United States of America, 99, 15898–15903.
Li, H. W., Zang, B.S., Deng, X. W., Wang, X. P. (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta. 234, 1007–1018.
Thomas, J. C., Sepahi, M., Arendall, B., & Bohnert, H. J. (1995). Enhancement of seed germination in high salinity by engineering mannitol expression in Arabidopsis thaliana. Plant, Cell and Environment, 18, 801–806.
Karakas, B., Ozias Akins, P., Stushnoff, C., Suefferheld, M., & Rieger, M. (1997). Salinity and drought tolerance of mannitol-accumulating transgenic tobacco. Plant, Cell and Environment, 20, 609–616.
Abebe, T., Guenzi, A. C., Martin, B., & Cushman, J. C. (2003). Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiology, 131, 1748–1755.
Molinari, H. B. C., Marur, C. J., Bespalhok, F. J. C., Kobayashi, A. K., Pileggi, M., Leite, R. P., Jr., et al. (2004). Osmotic adjustment in transgenic citrus rootstock Carrizo citrange (Citrus sinensis Osb × Poncirus trifoliate L. Raf) overproducing proline. Plant Science, 167, 1375–1381.
Zhu, X., Gong, H., Chen, G., Wang, S., & Zhang, C. (2005). Different solute levels in two spring wheat cultivars induced by progressive field water stress at different developmental stages. Journal of Arid Environments, 62, 1–14.
Kavi Kishore, P. B., Hong, Z., Miao, G. H., Hu, C. A., & Verma, D. P. S. (1995). Overexpression of pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiology, 108, 1387–1394.
Hong, Z., Lakkineni, K., Zhang, K., & Verma, D. P. S. (2000). Removal of feedback inhibition of 1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiology, 122, 1129–1136.
Zhu, B., Su, J., Chang, M. C., Verma, D. P. S., Fan, Y. L., & Wu, R. (1998). Overexpression of a pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water and salt stress in transgenic rice. Plant Science, 139, 41–48.
Su, J., & Wu, R. (2004). Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Science, 166, 941–948.
Nanjo, T., Kobayashi, M., Yoshiba, Y., Kakubari, Y., Yamaguchi-Shinozaki, K., & Shinozaki, K. (1999). Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Letters, 461, 205–210.
Ma, L., Zhou, E., Gao, L., Mao, X., Zhou, R., & Jia, J. (2008). Isolation, expression analysis and chromosomal location of P5CR gene in common wheat (Triticum aestivum L.). The South African Journal of Botany, 74, 705–712.
Rhodes, D., & Hanson, A. D. (1993). Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annual Review of Plant Physiology, 44, 357–384.
Holmstrom, K. O., Somersalo, S., Mandal, A., Palva, T. E., & Welin, B. (2000). Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. Journal of Experimental Botany, 51, 177–185.
Brouquisse, R., Weigel, P., Rhodes, D., Yocum, C. F., & Hanson, A. D. (1989). Evidence for a ferredoxin-dependent choline monooxygenase from spinach chloroplast stroma. Plant Physiology, 90, 322–329.
Weigel, P., Weretilnyk, E. A., & Hanson, A. D. (1986). Betaine aldehyde oxidation by spinach chloroplasts. Plant Physiology, 82, 753–759.
Lamark, T., Kaasen, I., Eshoo, M. W., Falkenberg, P., McDougall, J., & Strem, A. R. (1991). DNA sequence and analysis of bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Molecular Microbiology, 5, 1049–1064.
Bhattacharya, R. C., Maheswari, M., Dineshkumar, V., Kirti, P. B., Bhat, S. R., & Chopra, V. L. (2004). Transformation of Brassica oleracea var. capitata with bacterial betA gene enhances tolerance to salt stress. Scientia Horticulturae, 100, 215–227.
Quan, R., Shang, M., Zhang, H., Zhao, Y., & Zhang, J. (2004). Engineering of enhanced glycine betaine synthesis improves drought tolerance in maize. Plant Biotechnology Journal, 2, 477–486.
Cherian, S., Reddy, M. P., & Ferreira, R. B. (2006). Transgenic plants with improved dehydration-stress tolerance: Progress and future prospects. Biologia Plantarum, 50, 481–495.
Smirnoff, N. (1998). Plant resistance to environmental stress. Current Opinion in Biotechnology, 9, 214–219.
Bartels, D. (2001). Targeting detoxification pathways, an efficient approach to obtain plants with multiple stress tolerance. Trends in Plant Science, 6, 284–286.
Apel, K., & Hirt, H. (2004). Reactive oxygen species, metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology, 55, 373–399.
Miller, G., Suzuki, N., Yilmaz, S. C., & Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell and Environment, 33, 453–467.
Suzuki, N., Koussevitzky, S., Mittler, R., & Miller, G. (2011). ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell and Environment, 35, 259–270.
Ashraf, M. (2009). Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnology Advances, 27, 84–93.
Sun, W. H., Duan, M., Yang, D. F. S. S., & Meng, Q. W. (2010). Over-expression of StAPX in tobacco improves seed germination and increases early seedling tolerance to salinity and osmotic stresses. Plant Cell Reports, 29, 917–926.
Badawi, G. H., Kawano, N., Yamauchi, Y., Shimada, E., Sasaki, R., Kubo, A., et al. (2004). Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiologia Plantarum, 121, 231–238.
Lu, Z. Q., Liu, D. L., & Liu, S. K. (2007). Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Reports, 26, 1909–1917.
Kavitha, K., Venkataraman, G., & Parida, A. (2008). An oxidative and salinity stress induced peroxisomal ascorbate peroxidase from Avicennia marina, molecular and functional characterization. Plant Physiology and Biochemistry, 46, 794–804.
Sengupta, A., Heinen, J., Holaday, A. S., Barke, J. J., & Allan, R. D. (1993). Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proceedings of the National Academy of Sciences of the United States of America, 90, 1629–1633.
Prashanth, S. R., Sadhasivam, V., & Parida, A. (2008). Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Research, 17, 281–291.
Wang, Y., Ying, Y., Chen, J., & Wang, X. (2004). Transgenic Arabidopsis overexpressing Mn-SOD enhanced salt-tolerance. Plant Science, 167, 671–677.
Lee, Y. P., Kim, S. H., Bang, J. W., Lee, H. S., Kwak, S. S., & Kwon, S. Y. (2007). Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Reports, 26, 591–598.
Lee, Y. P., Baek, K. H., Lee, H. S., Kwak, S. S., Bang, J. W., & Kwon, S. Y. (2010). Tobacco seeds simultaneously over-expressing Cu/Zn superoxide dismutase and ascorbate peroxidase display enhanced seed longevity and germination rates under stress conditions. Journal of Experimental Botany, 61, 2499–2506.
Roxas, V. P., Smith, R. K., Jr., Allen, E. R., & Allen, R. D. (1997). Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nature Biotechnology, 15, 988–991.
Roxas, V. P., Lodhi, S. A., Garrett, D. K., Mahan, J. R., & Allen, R. D. (2000). Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant and Cell Physiology, 41, 1229–1234.
Igarashi, K., & Kashiwagi, K. (2000). Polyamines, mysterious modulators of cellular functions. Biochemical and Biophysical Research Communications, 271, 559–564.
Alcazar, R., Altabella, T., Bortolotti, F. M. C., Koncz, M. R. C., Carrasco, P., & Tiburcio, A. F. (2010). Polyamines, molecules with regulatory functions in plant abiotic stress tolerance. Planta, 231, 1237–1249.
Gill, S., & Tuteja, N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signaling and Behavior, 5, 26–33.
Minocha, S. C., & Sun, D. Y. (1997). Stress tolerance in plants through transgenic manipulation of polyamine biosynthesis. Plant Physiology Supplement, 114, 297 (Abstract #1552).
Kumria, R., & Rajam, M. V. (2002). Ornithine decarboxylase transgene in tobacco affects polyamines, in vitro morphogenesis and response to salt stress. Journal of Plant Physiology, 159, 983–990.
Roy, M., & Wu, R. (2001). Arginine decarboxylase transgene expression and analysis of environmental stress tolerance in transgenic rice. Plant Science, 160, 869–875.
Waie, B., & Rajam, M. V. (2003). Effect of increased polyamine biosynthesis on stress responses in transgenic tobacco by introduction of human S-adenosylmethionine gene. Plant Science, 164, 727–734.
Shi, H., Ishitani, M., Kim, C., & Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences of the United States of America, 97, 6896–6901.
Shi, H., Lee, B. H., Wu, S. J., & Zhu, J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology, 21, 81–85.
Gaxiola, R. A., Rao, R., Sherman, A., Grifasi, P., Alpier, S. L., & Fink, G. R. (1999). The Arabidopsis thaliana proton transporters, AtNHX1 and Avp1, can function in cation detoxification in yeast. Proceedings of the National Academy of Sciences of the United States of America, 96, 1480–1485.
Apse, M. P., Aharon, G. S., Snedden, W. A., & Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science, 285, 1256–1258.
Chauhan, S., Forsthoefel, N., Ran, Y., Quigley, F., Nelson, D. E., & Bohnert, H. J. (2000). Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. The Plant Journal, 24, 511–522.
Hamada, A., Shono, M., **a, T., Ohta, M., Hayashi, Y., Tanaka, A., et al. (2001). Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Molecular Biology, 46, 35–42.
Ma, X. L., Zhang, Q., Shi, H. Z., Zhu, J. K., Zhao, Y. X., Ma, C. L., et al. (2004). Molecular cloning and different expression of a vacuolar Na+/H+ antiporter gene in Suaeda salsa under salt stress. Biologia Plantarum, 48, 219–225.
**a, T., Apse, M. P., Aharon, G. S., & Blumwald, E. (2002). Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiologia Plantarum, 116, 206–212.
Jha, A., Joshi, M., Yadav, N. S., Agarwal, P. K., & Jha, B. (2010). Cloning and characterization of the Salicornia brachiata Na+/H+ antiporter gene SbNHX1 and its expression by abiotic stress. Molecular Biology Reports, 38, 1965–1973.
Bordas, M., Montesinos, C., Dabauza, M., Salvador, A., Roig, L. A., Serrano, R., et al. (1997). Transfer of the yeast salt tolerance gene HAL1 to Cucumis melo L. Cultivars and in vitro evaluation of salt tolerance. Transgenic Research, 6, 41–50.
Gisbert, C., Rus, A. M., Bolarin, M. C., Lopez-Coronado, M., Arrillaga, I., Montesinos, C., et al. (2000). The yeast HAL1 gene improves salt tolerance of transgenic tomato. Plant Physiology, 123, 393–402.
Shi, H., **ong, L., Stevenson, B., Lu, T., & Zhu, J. K. (2002). The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. The Plant Cell, 14, 575–588.
Qiu, Q. S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., & Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly sensitive (SOS) pathway. Journal of Biological Chemistry, 279, 207–215.
Gao, X. Z., Zhao, R. Y., & Zhang, H. (2003). Overexpression of SOD2 increases salt tolerance of Arabidopsis. Plant Physiology, 133, 1873–1881.
Wu, L., Fan, Z., Guo, L., Li, Y., Chen, Z. L., & Qu, L. J. (2005). Overexpression of the bacterial nhaA gene in rice enhances salt and drought tolerance. Plant Science, 168, 297–302.
Oh, D. H., Leidi, E., Zhang, Q., Hwang, S. M., Li, Y., Quintero, F. J., et al. (2009). Loss of halophytism by interference with SOS1 expression. Plant Physiology, 151, 210–222.
Yang, Q., Chen, Z. Z., Zhou, X. F., Yin, H. B., Li, X., **n, X. F., et al. (2009). Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant, 2, 22–31.
Brini, F., Hanin, M., Mezghani, I., Berkowitz, G. A., & Masmoudi, K. (2007). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt-and drought-stress tolerance in Arabidopsis thaliana plants. Journal of Experimental Botany, 58, 301–308.
Serrano, R., Mulet, J. M., Rios, G., Marquez, J. A., de Larrinoa, I. F., Leube, M. P., et al. (1999). A glimpse of the mechanisms of ion homeostasis during salt stress. Journal of Experimental Botany, 50, 1023–1036.
Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., Alper, S. L., et al. (2001). Drought-and salt-tolerant plants result from overexpression of the AVP1 H+ pump. Proceedings of the National Academy of Sciences of the United States of America, 98, 11444–11449.
Deswal, R., Chakravarty, T. N., & Sopory, S. K. (1993). The glyoxalase system in higher plants, regulation in growth and differentiation. Biochemical Society Transactions, 21, 527–530.
Paulus, C., Knollner, B., & Jacobson, H. (1993). Physiological and biochemical characterization of glyoxalase I, a general marker for cell proliferation, from a soybean cell suspension. Planta, 189, 561–566.
Espartero, J., Sanchez-Aguayo, I., & Pardo, J. M. (1995). Molecular characterization of glyoxalase I from a higher plant, upregulation by stress. Plant Molecular Biology, 29, 1223–1233.
Veena, Reddy, V. S., & Sopory, S. K. (1999). Glyoxalase I from Brassica juncea, molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. The Plant Journal, 17, 385–395.
Jain, M., Choudhary, D., Kale, R. K., & Bhalla-Sarin, N. (2002). Salt-and glyphosate-induced increase in glyoxalase I activity in cell lines of groundnut (Arachis hypogaea). Physiologia Plantarum, 114, 499–505.
Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proceedings of the National Academy of Sciences of the United States of America, 100, 14672–14677.
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., & Sopory, S. K. (2008). Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Research, 17, 171–180.
Sunkar, R., Chinnusamy, V., Zhu, J., & Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science, 12, 301–309.
Sunkar, R., & Zhu, J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell, 16, 2001–2019.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
Jones-Rhoades, M. W., & Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Molecular Cell, 14, 787–799.
Lu, S., Sun, Y. H., Shi, R., Clark, C., Li, L., & Chiang, V. L. (2005). Novel and mechanical stress responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. The Plant Cell, 17, 2186–2203.
Shukla, L. I., Chinnusamy, V., & Sunkar, R. (2008). The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochimica et Biophysica Acta, 1779, 743–748.
Sunkar, R., Kapoor, A., & Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell, 18, 2415.
Zhao, B., Liang, R., Ge, L., Li, W., **ao, H., Lin, H., et al. (2007). Identification of drought-induced microRNAs in rice. Biochemical and Biophysical Research Communications, 354, 585–590.
Kulcheski, F. R., de Oliveira, L. F., Molina, L. G., Almerao, M. P., Rodrigues, F. A., Marcolino, J., et al. (2011). Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics, 12, 307.
Covarrubias, A. A., & Reyes, J. L. (2009). Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant, Cell and Environment, 33, 481–489.
Sunkar, R. (2010). MicroRNAs with macro-effects on plant stress responses. Seminars in Cell & Developmental Biology, 2, 1805–1811.
Lu, S., Sun, Y. H., & Chiang, V. L. (2008). Stress-responsive microRNAs in Populus. The Plant Journal, 55, 131–151.
Zhao, B. T., Ge, L. F., Liang, R. Q., Li, W., Ruan, K. C., Lin, H. X., et al. (2009). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Molecular Biology, 10, 29.
Feng, X. M., You, C. X., Qiao, Y., Mao, K., & Hao, Y. J. (2010). Ectopic overexpression of Arabidopsis AtmiR393a gene changes auxin sensitivity and enhances salt resistance in tobacco. Acta Physiologia Plantarum, 32, 997–1003.
Gao, P., Bai, X., Yang, L., Lv, D., Li, Y., Cai, H., et al. (2010). Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta, 231, 991–1001.
Gao, P., Bai, X., Yang, L., Lv, D., Pan, X., Li, Y., et al. (2011). osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Molecular Biology Reports, 38, 237–242.
**a, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE. doi:10.1371/journal.pone.0030039.
Liu, D., Song, Y., Chen, Z., & Yu, D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum, 136(2), 223–236.
Wang, F., Liu, R., Wu, G., Lang, C., Chen, J., & Shi, C. (2012). Specific downregulation of the bacterial-type PEPC gene by artificial MicroRNA improves salt tolerance in Arabidopsis. Plant Molecular Biology Reporter. doi:10.1007/s11105-012-0418-6.
Kim, J. Y., Lee, H. J., Jung, H. J., Maruyama, K., Suzuki, N., & Kang, H. (2010). Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta, 232, 1447–1454.
Xue, Z. Y., Zhi, D. Y., Xue, G. P., Zhang, H., Zhao, Y. H., & **a, G. M. (2004). Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Science, 167, 849–859.
Waterer, D., Benning, N. T., Wu, G., Luo, X., Liu, X., Gusta, M., et al. (2010). Evaluation of abiotic stress tolerance of genetically modified potatoes (Solanum tuberosum cv. Desiree). Molecular Breeding, 25, 527–540.
Cheong, Y. H., Sung, S. J., Kim, B. Gi., Pandey, G. K., Cho, J. S., Kim, K. N., et al. (2010). Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Molecules and Cells, 29, 159–165.
**ng, Y., Jia, W., & Zhang, J. (2007). AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. Journal of Experimental Botany, 58, 2969–2981.
Ning, J., Li, X., Hicks, L. M., & **ong, L. (2010). A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology, 152, 876–890.
**ang, Y., Huang, Y., & **ong, L. (2007). Characterization of stress responsive CIPK genes in rice for stress tolerance improvement. Plant Physiology, 144, 1416–1428.
Xu, J., Tian, Y. S., Peng, R. H., **ong, A. S., Zhu, B., **, X. F., et al. (2010). AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta, 231, 1251–1260.
Kovtun, Y., Chiu, W. L., Tena, G., & Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proceedings of the National Academy of Sciences of the United States of America, 97, 2940–2945.
Shou, H., Bordallo, P., & Wang, K. (2004). Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. Journal of Experimental Botany, 55, 1013–1019.
Ying, S., Zhang, D. F. Li, H. Y., Liu, Y. H., Shi, Y. S., Song, Y. C., et al. (2011). Isolation and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Reports. doi:10.1007/s00299-011-1077-z.
Zhang, L., **, D., Li, S., Gao, Z., Zhao, S., Shi, J., et al. (2011) A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Molecular Biology. doi:10.1007/s11103-011-9788-7.
Xu, G. Y., Rocha, P. S. C. F., Wang, M. L., Xu, M. L., Cui, Y. C., Li, L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta, 234, 47–59. doi:10.1007/s00425-011-1386-z.
Zhang, H., Mao, X., Wang, C. & **g, R. (2010). Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLOS ONE. doi:10.1371/journal.pone.0016041.g006.
James, V. A., Neibaur, I., & Altpeter, F. (2008). Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L. in turf and forage grass (Paspalum notatum Flugge) enhances abiotic stress tolerance. Transgenic Research, 17, 93–104.
Liu, N., Zhong, N. Q., Wang, G. L., Li, L. J., Liu, X. L., He, Y. K., et al. (2007). Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta, 226, 827–838.
Bhatnagar, P., Devi, M. J., Reddy, D. S., Lavanya, M., Vadez, V., Serraj, R., et al. (2007). Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Reports, 26, 2071–2082.
Xu, Z. S., **a, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y., Li, L. C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Molecular Biology, 65, 719–732.
Zhao, J. S., Ren, W., Zhi, D., Wang, L., & **a, G. (2007). Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant Cell Reports, 26, 1521–1528.
Tong, Z., Hong, B., Yang, Y., Li, Q., Ma, N., Ma, C., et al. (2009). Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Molecular Biology, 71, 115–129.
Xu, Z. S., Ni, Z. Y., Li, Z. Y., Li, L. C., Chen, M., Gao, D. Y., et al. (2009). Isolation and functional characterization of HvDREB1, a gene encoding a dehydration-responsive element binding protein in Hordeum vulgare. Journal of Plant Research, 122, 121–130.
Chen, J., **a, X., & Yin, W. (2009). Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica. Biochemical and Biophysical Research Communications, 378, 483–487.
Agarwal, P., Agarwal, P. K., Joshi, A. J., Reddy, M. K., & Sopory, S. K. (2010). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Molecular Biology Reports, 37, 1125–1135.
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Molecular Genetics and Genomics, 283, 185–196.
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S. P., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stress in Zea mays L. The Plant Journal, 50, 54–59.
Chen, M., Wang, Q. Y., Cheng, X. G., Xu, Z. S., Li, L. C., Ye, X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochemical and Biophysical Research Communications, 353, 299–305.
Gao, S. Q., Chen, M., **a, L. Q., **u, H. J., Xu, Z. S., Li, L. C., et al. (2009). A cotton (Gossypium hirsutum) DREB-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Reports, 28, 301–311.
Zhang, Y., Chen, C., **, X. F., **ong, A. S., Peng, R. H., Hong, Y. H., et al. (2009). Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. Biochemistry and Molecular Biology Reports, 42, 486–492.
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proceedings of the National Academy of Sciences of the United States of America, 103, 18822–18827.
Wang, Q., Guan, Y., Wu, Y., Chen, H., Chen, F., & Chu, C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Molecular Biology, 67, 589–602.
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnology Journal, 9, 230–249.
**anjun, P., **ngyong, M., Weihong, F., Man, S., Liqin, C., Alam, I., et al. (2011). Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymus chinensis. Plant Cell Reports, 30, 1493–1502.
Li, D., Zhang, Y., Hu, X., Shen, X., Ma, L., Su, Z., et al. (2011). Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biology, 11, 109.
Siddiqua, M., & Nassuth, A. (2011). Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant, Cell and Environment. doi:10.1111/j.1365-3040.2011.02334.x.
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. Journal of Experimental Botany, 60, 3781–3796.
Villalobos, M. A., Bartels, D., & Iturriaga, G. (2004). Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiology, 135, 309–324.
Park, M. Y., Kang, J. Y., & Kim, S. Y. (2011). Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Molecules and Cells, 31, 447–454. doi:10.1007/s10059-011-0300-7.
Kim, S. G., Lee, A. K., Yoon, H. K., & Park, C. M. (2008). A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. The Plant Journal, 55, 77–88.
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., & McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Molecular Plant, 1–16.
Majee, M., Maitra, S., Dastidar, K. G., Pattnaik, S., Chatterjee, A., Hait, N. C., et al. (2004). A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice, molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. Journal of Biological Chemistry, 279, 28539–28552.
Tang, W., Peng, X., & Newton, R. J. (2005). Enhanced tolerance to salt stress in transgenic loblolly pine simultaneously expressing two genes encoding mannitol-1-phosphate dehydrogenase and glucitol-6-phosphate dehydrogenase. Plant Physiology and Biochemistry, 43, 139–146.
Sawahel, W. A., & Hassan, A. H. (2002). Generation of transgenic wheat plants producing high levels of the osmoprotectant proline. Biotechnology Letters, 24, 721–725.
Hmida-Sayari, A., Gargouri-Bouzid, R., Bidani, A., Jaoua, L., Savoure, A., & Jaoua, S. (2005). Overexpression of Δ1–pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Science, 169, 746–752.
Hayashi, H., Alia, L., Mustardy, P., Deshnium, M. I., & Murata, N. (1997). Transformation of Arabidopsis thaliana with the codA gene for choline oxidase; accumulation of glycine betaine and enhanced tolerance to salt and cold stress. Plant J., 12, 133–142.
Sakamoto, A., Alia, H., & Murata, N. (1998). Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Molecular Biology, 38, 1011–1019.
Mohanty, A., Kathuria, H., Ferjani, A., Sakamoto, A., Mohanty, P., Murata, N., et al. (2002). Transgenics of an elite indica rice variety Pusa Basmati 1 harbouring the codA gene are highly tolerant to salt stress. TAG. Theoretical and Applied Genetics, 106, 51–57.
Prasad, K. V. S. K., Sharmila, P., Kumar, P. A., & Saradhi, P. P. (2000). Transformation of Brassica juncea (L.) Czern with bacterial cod.A gene enhances its tolerance to salt stress. Molecular Breeding, 6, 489–499.
Guo, B. H., Zhang, Y. M., Li, H. J., Du, L. Q., Li, Y. X., Zhang, J. S., et al. (2000). Transformation of wheat with a gene encoding for the betaine aldehyde dehydrogenase (BADH). Acta Botanica Sinica, 42, 279–283.
Alet, A. I., Sanchez, D. H., Cuevas, J. C., Valle, S. D., Altabella, T., Tiburcio, A. F., et al. (2011). Putrescine accumulation in Arabidopsis thaliana transgenic lines enhances tolerance to dehydration and freezing stress. Plant Signaling and Behavior, 6, 278–286.
Wang, Y., Wisniewski, M. E., Meilan, R., Webb, R., Fuchigami, L., & Boyer, C. (2005). Overexpression of cytosolic ascorbate peroxidase in tomato (Lycopersicon esculentum) confers tolerance to chilling and salt stress. Journal of the American Society for Horticultural Science, 130, 167–173.
Qi, Y. C., Liu, W. Q., Qiu, L. Y., Zhang, S. M., Ma, L., & Zhang, H. (2010). Overexpression of glutathione S-transferase gene increases salt tolerance of Arabidopsis. The Russian Journal of Plant Physiology, 57, 233–240.
Wang, F. Z., Wang, Q. B., Kwon, S. Y., Kwak, S. S., & Su, W. A. (2005). Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. Journal of Plant Physiology, 162, 465–472.
Chatzidimitriadou, K., Nianiou-Obeidat, I., Madesis, P., Perl-Treves, R., & Tsaftaris, A. (2009). Expression of SOD transgene in pepper confer stress tolerance and improve shoot regeneration. Electronic Journal of Biotechnology, 12, 4.
Ushimaru, T., Nakagawa, T., Fujioka, Y., Daicho, K., Naito, M., Yamauchi, Y., et al. (2006). Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. Journal of Plant Physiology, 163, 1179–1184.
Eltayeb, A. E., Kawano, N., Badawi, G. H., Kaminaka, H., Sanekata, T., Shibahara, T., et al. (2007). Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta, 225, 1255–1264.
Kavitha, K., George, S., Venkataraman, G., & Parida, A. (2010). A salt-inducible chloroplastic monodehydroascorbate reductase from halophyte Avicennia marina confers salt stress tolerance on transgenic plants. Biochimie, 92, 1321–1329.
Al-Taweel, K., Iwaki, T., Yabuta, Y., Shigeoka, S., Murata, N., & Wadano, A. (2007). A bacterial transgene for catalase protects translation of d1 protein during exposure of salt-stressed tobacco leaves to strong light. Plant Physiology, 145, 258–265.
Moriwaki, T., Yamamoto, Y., Aida, T., Funahashi, T., Shishido, T., Asada, M., et al. (2008). Overexpression of the Escherichia coli catalase gene, katE, enhances tolerance to salinity stress in the transgenic indica rice cultivar, BR5. Plant Biotechnology Reports, 2, 41–46.
Capell, T., Bassie, L., & Christou, P. (2004). Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proceedings of the National Academy of Sciences of the United States of America, 101, 9909–9914.
Alcázar, R., Planas, J., Saxena, T., Zarza, X., Bortolotti, C., Cuevas, J. C., et al. (2010). Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants overexpressing the homologous Arginine decarboxylase 2 gene. Plant Physiology and Biochemistry, 48, 547–552.
Roy, M., & Wu, R. (2002). Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Science, 163, 987–992.
Kasukabe, Y., He, L., Nada, K., Misawa, S., Ihara, I., & Tachibana, S. (2004). Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant and Cell Physiology, 45, 712–722.
Wen, X. P., Pang, X. M., Matsuda, N., Kita, M., Inoue, H., Hao, Y. J., et al. (2008). Over-expression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Research, 17, 251–263.
Zhang, H. X., & Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nature Biotechnology, 19, 765–768.
Zhang, H. X., Hodson, J. N., Williams, J. P., & Blumwald, E. (2001). Engineering salt tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proceedings of the National Academy of Sciences of the United States of America, 98, 12832–12836.
Zhao, J. S., Zhi, D. Y., & Xue, Z. Y. (2007). Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+/H+ antiporter gene from Arabidopsis. Journal of Plant Physiology, 164, 1377–1383.
Chen, L. H., Zhang, B., & Xu, Z. Q. (2008). Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Research, 17, 121–132.
Liu, H., Wang, Q., Yu, M., Zhang, Y., Wu, Y., & Zhang, H. (2008). Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively overexpressing an Arabidopsis thaliana vacuolar Na+/H+ antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage root. Plant, Cell and Environment, 31, 1325–1334.
Tian, N., Wang, J., & Xu, Z. Q. (2010). Overexpression of Na+/H+ antiporter gene AtNHX1 from Arabidopsis thaliana improves the salt tolerance of kiwifruit (Actinidia deliciosa). South African Journal of Botany, 77, 160–169.
**ao-Yan, Y., Ai-Fang, Y., Ke-Wei, Z., & Ju-Ren, Z. (2004). Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1; Gene. Acta Botanica Sinica, 46, 854–861.
Ohta, M., Hayashi, Y., Nakashima, A., Hamada, A., Tanaka, A., Nakamura, T., et al. (2002). Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Letters, 532, 279–282.
Fukuda, A., Nakamura, A., Tagiri, A., Tanaka, H., Miyao, A., Hirochika, H., et al. (2004). Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant and Cell Physiology, 45, 146–159.
Wu, C. A., Yang, G. D., Meng, Q. W., & Zheng, C. C. (2004). The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant and Cell Physiology, 45, 600–607.
Lu, S. Y., **g, Y. X., Shen, S. H., Zhao, H. Y., Ma, L. Q., Zhou, X. J., et al. (2005). Antiporter Gene from Hordeum brevisubulatum (Trin.) link and Its overexpression in transgenic tobaccos. Journal of Integrative Plant Biology, 47, 343–349.
Rajagopal, D., Agarwal, P., Tyagi, W., Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2007). Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Molecular Breeding, 19, 137–151.
Verma, D., Singla-Pareek, S. L., Rajagopal, D., Reddy, M. K., & Sopory, S. K. (2007). Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. Journal of Biosciences, 32, 621–626.
Qiao, W. H., Zhao, X. Y., & Li, W. (2007). Overexpression of AeNHX1, a root-specific vacuolar Na+/H+ antiporter from Agropyron elongatum, confers salt tolerance to Arabidopsis and Festuca plants. Plant Cell Reports, 26, 1663–1672.
Zhang, G. H., Su, Q., An, L. J., & Wu, S. (2008). Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiology and Biochemistry, 46, 117–126.
Li, W., Wang, D., **, T., Chang, Q., Yin, D., Xu, S., et al. (2010). The vacuolar Na+/H+ antiporter gene SsNHX1from the halophyte Salsola soda confers salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Molecular Biology Reporter, 29, 278–290.
Li, Y., Zang, Y., Feng, F., Liang, D., Cheng, L., & Shi, S. M. (2010). Overexpression of a Malus vacuolar Na+/H+ antiporter gene (MdNHX1) in apple rootstock M.26 and its influence on salt tolerance. Plant Cell, Tissue and Organ Culture, 102, 337–345.
Wu, C., Gao, X., Kong, X., Zhao, Y., & Zhang, H. (2009). Molecular cloning and functional analysis of a Na+/H+ antiporter gene ThNHX1 from a halophytic plant Thellungiella halophila. Plant Molecular Biology Reporter, 27, 1–12.
Guan, B., Hu, Y., Zeng, Y., Wang, Y., & Zhang, F. (2011). Molecular characterization and functional analysis of a vacuolar Na+/H+ antiporter gene (HcNHX1) from Halostachys caspica. Molecular Biology Reports, 38, 1889–1899.
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Molecular Biotechnology. doi:10.1007/s12033-011-9399-1.
Wei, Q., Guo, Y. J., Cao, H. M., & Kuai, B. K. (2011). Cloning and characterization of an AtNHX2-like Na+/H+ antiporter gene from Ammopiptanthus mongolicus (Leguminosae) and its ectopic expression enhanced drought and salt tolerance in Arabidopsis thaliana. Plant Cell, Tissue and Organ Culture, 105, 309–316.
Bayat, F., Shiran, B., Belyaev, D. V., Yur’eva, N. O., Sobol’kova, G. I., Alizadeh, H., et al. (2010). Potato plants bearing a vacuolar Na+/H+ antiporter HvNHX2 from barley are characterized by improved salt tolerance. Russian Journal of Plant Physiology, 57, 696–706.
Zhao, F. Y., Guo, S. L., Zhang, H., & Zhao, Y. X. (2006). Expression of yeast SOD2 in transgenic rice results in increased salt tolerance. Plant Science, 170, 216–224.
Martínez-Atienza, J., Jiang, X. Y., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiology, 143, 1001–1012.
Wu, Y., Ding, N., Zhao, X., Zhao, M., Chang, Z., Liu, J., et al. (2007). Molecular characterization of PeSOS1, the putative Na+/H+ antiporter of Populus euphratica. Plant Molecular Biology, 65, 1–11.
Olías, R., Eljakaoui, Z., Li, J., Morales, P. A. D., Marín-manzano, M. C., Pardo, J. M., et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant, Cell and Environment, 32, 904–916.
D’yakova, E. V., Rakitin, A. L., Kamionskaya, A. M., Baikov, A. A., Lahti, R., Ravin, N. V., et al. (2006). A study of the effect of expression of the gene encoding the membrane H+-pyrophosphatase of Rhodospirillum rubrum on salt resistance of transgenic tobacco plants. Doklady Biological Sciences, 409, 346–348.
Gao, F., Gao, Q., Duan, X. G., Yue, G. D., Yang, A. F., & Zhang, J. R. (2006). Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. Journal of Experimental Botany, 57, 3259–3270.
Li, B., Wei, A. Y., Song, C. X., Li, N., & Zhang, J. R. (2008). Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnology Journal, 6, 146–159.
Lv, S. L., Zhang, K. W., Gao, Q., Lian, L. J., Song, Y. J., & Zhang, J. R. (2008). Overexpression of an H + -PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant and Cell Physiology, 49, 1150–1164.
Bao, A. K., Wang, S. M., Wu, G. Q., **, J. J., Zhang, J. L., & Wang, C. M. (2009). Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Science, 176, 232–240.
Zhao, F. Y., Zhang, X. J., Li, P. H., Zhao, Y. X., & Zhang, H. (2006). Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1. Mol. Breed., 17, 341–353.
Guo, S., Yin, H., Zhang, X., Zhao, F., Li, P., Chen, S., et al. (2006). Molecular cloning and characterization of a vacuolar H+-pyrophopsatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Molecular Biology, 60, 41–50.
Pasapula, V., Shen, G., Kuppu, S., Paez-Valencia, J., Mendoza, M., Hou, P., et al. (2010). Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnology Journal, 9, 88–99.
Li, Z., Baldwin, C. M., Hu, Q., Liu, H., & Luo, H. (2010). Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic cree** bentgrass (Agrostis stolonifera L.). Plant, Cell and Environment, 33, 272–289.
Lv, S. L., Lian, L. J., Tao, P. L., Li, Z. X., Zhang, K. W., & Zhang, J. R. (2009). Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta, 229, 899–910.
Jacobs, A., Ford, K., Kretschmer, J., & Tester, M. (2011). Rice plants expressing the moss sodium pum** ATPase PpENA1 maintain greater biomass production under salt stress. Plant Biotechnology Journal, 9, 838–847.
Gao, C., Wang, Y., Jiang, B., Liu, G., Yu, L., Wei, Z., et al. (2011). A novel vacuolar membrane H+-ATPase c subunit gene (ThVHAc1) from Tamarix hispida confers tolerance to several abiotic stresses in Saccharomyces cerevisiae. Molecular Biology Reports, 38, 957–963.
Acknowledgments
The financial support received from CSIR and DST New Delhi is gratefully acknowledged. Kapil Gupta acknowledges the award of CSIR-SRF.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Agarwal, P.K., Shukla, P.S., Gupta, K. et al. Bioengineering for Salinity Tolerance in Plants: State of the Art. Mol Biotechnol 54, 102–123 (2013). https://doi.org/10.1007/s12033-012-9538-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9538-3