Abstract
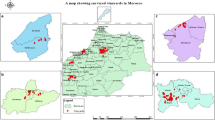
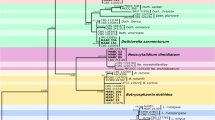
Grapevine production is economically indispensable for the global wine industry. Currently, Mexico cultivates grapevines across approximately 28 500 hectares, ranking as the 26th largest producer worldwide. Given its significance, early detection of plant diseases’ causal agents is crucial for preventing outbreaks. Consequently, our study aimed to identify fungal strains in grapevines exhibiting trunk disease symptoms and assess their enzymatic capabilities as indicators of their phytopathogenic potential. We collected plant cultivars, including Malbec, Shiraz, and Tempranillo, from Querétaro, Mexico. In the laboratory, we superficially removed the plant bark to prevent external contamination. Subsequently, the sample was superficially disinfected, and sawdust was generated from the symptomatic tissue. Cultivable fungal strains were isolated using aseptic techniques from the recovered sawdust. Colonies were grown on PDA and identified through a combination of microscopy and DNA-sequencing of the ITS and LSU nrDNA regions, coupled with a BLASTn search in the GenBank database. We evaluated the strains’ qualitative ability to degrade cellulose, starch, and lignin using specific media and stains. Using culture morphology and DNA-sequencing, 13 species in seven genera were determined: Acremonium, Aspergillus, Cladosporium, Dydimella, Fusarium, Sarocladium, and Quambalaria. Some isolated strains were able to degrade cellulose or lignin, or starch. These results constitute the first report of these species community in the Americas. Using culture-dependent and DNA-sequencing tools allows the detection of fungal strains to continue monitoring for early prevention of the GTD.





Similar content being viewed by others

References
INEGI (2019) Encuesta Nacional Agropecuaria 2019. https://www.inegi.org.mx/contenidos/programas/ena/2019/tabulados/ena19_nac_agri01.xlsx. Accessed 5 Mar 2023
FAO (2020) Crops and livestock products. In: FAOSTAT. https://www.fao.org/faostat/en/#data/QCL Accessed 5 Mar 2023
de Almeida AB, Concas J, Campos MD et al (2020) Endophytic fungi as potential biological control agents against grapevine trunk diseases in alentejo region. Biology (Basel) 9:1–23. https://doi.org/10.3390/biology9120420
Hrycan J, Hart M, Bowen P et al (2020) Grapevine trunk disease fungi: their roles as latent pathogens and stress factors that favour disease development and symptom expression. Phytopathol Mediterr 59:395–424. https://doi.org/10.14601/Phyto-11275
Niem JM, Billones-Baaijens R, Stodart B, Savocchia S (2020) Diversity profiling of grapevine microbial endosphere and antagonistic potential of endophytic pseudomonas against grapevine trunk diseases. Front Microbiol 11:477. https://doi.org/10.3389/fmicb.2020.00477
Maldonado-González MM, Sodupe MA, Vicente CB, Muñoz RB et al (2018) Enfermedades fúngicas de la madera de la vid: líneas de investigación actuales y últimos avances para su control. Cuad campo 61:28–35
Rangel-Montoya EA, Rolshausen PE, Hernandez-Martinez R (2023) Unravelling the colonization mechanism of Lasiodiplodia brasiliensis in grapevine plants. Phytopathol Mediterr 62:135–149. https://doi.org/10.36253/phyto-14198
Claverie M, Notaro M, Fontaine F, Wery J (2020) Current knowledge on Grapevine Trunk Diseases with complex etiology: a systemic approach. Phytopathol Mediterr 59:29–53
Fischer M (2019) Grapevine trunk diseases in German viticulture. III. Biodiversity and spatial distribution of fungal pathogens in rootstock mother plants and possible relation to leaf symptoms. Vitis 58:141–149. https://doi.org/10.5073/vitis.2019.58.141-149
Narmani A, Arzanlou M (2019) Quambalaria cyanescens, a new fungal trunk pathogen associated with grapevine decline in Iran. Crop Prot 124:104875. https://doi.org/10.1016/j.cropro.2019.104875
Bustamante MI, Elfar K, Smith RJ et al (2022) First Report of Fusarium annulatum associated with young vine decline in California. Plant Dis 106:2752
Winkler AJ (1974) General viticulture. Univ of California Press, Oakland
Aguilar-Sánchez G (2020) Diferenciación de tierras agrícolas en el municipio de Tequisquiapan, Querétaro. Revista Geográfica de América Central 2:121–143. https://doi.org/10.15359/rgac.65-2.5
Barnett HL, Hunter BB (1998) Illustrated genera of imperfect fungi. The American phytopathological society. US Dep Agric Agric Res Serv Washingt State Univ Pullman APS Press USA St Paul, Minnesota USA
Vogel HJ (1956) A convenient growth medium for neurospora crassa. Microb Genet Bull 13:42–47
Riddell RW (1950) Permanent stained mycological preparations obtained by slide culture. Mycologia 42:265–270
Kiffer E, Michel M (2000) The Deuteromycetes, mitosporic fungi: classification and generic keys. CRC Press, Boca Raton
Ángeles-Argáiz RE, Flores-García A, Ulloa M et al (2016) Commercial Sphagnum peat moss is a vector for exotic ectomycorrhizal mushrooms. Biol Invasions 18:89–101. https://doi.org/10.1007/s10530-015-0992-2
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Hopple JS, Vilgalys R (1999) Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol Phylogenet Evol 13(1):1–19. https://doi.org/10.1006/mpev.1999.0634
Shahriarinour M, Wahab MNA, Ariff A, Mohamad R (2011) Screening, isolation and selection of cellulolytic fungi from oil palm empty fruit bunch fibre. Biotechnology 10:108–113. https://doi.org/10.3923/biotech.2011.108.113
Pointing SB (1999) Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers 2:17–33
Sujeeta KM, Shikha M, Khushboo S (2017) Isolation and screening of amylase producing fungi. Int J Curr Microbiol Appl Sci 6:783–788. https://doi.org/10.20546/ijcmas.2017.604.098
Batista-García RA, Balcázar-López E, Miranda-Miranda E et al (2014) Characterization of lignocellulolytic activities from a moderate halophile strain of Aspergillus caesiellus isolated from a sugarcane bagasse fermentation. PLoS ONE 9:e105893. https://doi.org/10.1371/journal.pone.0105893
Hammer O, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Arzanlou M, Moshari S, Salari M, Badali H (2013) Molecular characterization and pathogenicity of Phaeoacremonium spp. associated with esca disease of grapevine in Northern Iran. Arch Phytopathol Plant Prot 46:375–388. https://doi.org/10.1080/03235408.2012.741443
De Beer ZW, Begerow D, Bauer R et al (2006) Phytogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Stud Mycol 55:289–298. https://doi.org/10.3114/sim.55.1.289
Chen AJ, Frisvad JC, Sun BD et al (2016) Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Stud Mycol 84:1–118. https://doi.org/10.1016/j.simyco.2016.10.001
Bruez E, Vallance J, Gerbore J et al (2014) Analyses of the temporal dynamics of fungal communities colonizing the healthy wood tissues of esca leaf-symptomatic and asymptomatic vines. PLoS ONE 9:e95928. https://doi.org/10.1371/journal.pone.0095928
Silva-Valderrama I, Toapanta D, de los MMA et al (2021) Biocontrol potential of grapevine endophytic and rhizospheric fungi against trunk pathogens. Front Microbiol 11:1–13. https://doi.org/10.3389/fmicb.2020.614620
Ferrigo D, Causin R, Raiola A (2017) Effect of potential biocontrol agents selected among grapevine endophytes and commercial products on crown gall disease. Biocontrol 62:821–833. https://doi.org/10.1007/s10526-017-9847-3
Park SW, Nguyen TTT, Lee HB (2017) Characterization of two species of Acremonium (unrecorded in Korea) from soil samples: Acremonium variecolor and Acremonium persicinumw. Mycobiology 45:353–361. https://doi.org/10.5941/MYCO.2017.45.4.353
Deb D, Khan A, Dey N (2020) Phoma diseases: epidemiology and control. Plant Pathol 69:1203–1217. https://doi.org/10.1111/ppa.13221
Cimmino A, Bahmani Z, Masi M et al (2022) Phytotoxins produced by Didymella glomerata and Truncatella angustata, associated with grapevine trunk diseases (GTDs) in Iran. Nat Prod Res 36:4322–4329. https://doi.org/10.1080/14786419.2021.1979544
González V, Tello ML (2011) The endophytic mycota associated with Vitis vinifera in central Spain. Fungal Divers 47:29–42. https://doi.org/10.1007/s13225-010-0073-x
Dissanayake AJ, Purahong W, Wubet T et al (2018) Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers 90:85–107. https://doi.org/10.1007/s13225-018-0399-3
Machowicz-Stefanik Z, Król E (2013) Characterization of Phoma negriana, a new species from grapevine canes in Poland. Acta Mycol 42:113–117. https://doi.org/10.5586/am.2007.011
Gonzalez-Perez E, Yanez-Morales MJ, Ortega-Escobar HM, Velazquez-Mendoza J (2008) First report of Acremonium strictum and Gliocladium roseum causing basal stem and corm rot of Gladiolus grandiflorus in Mexico. J Plant Pathol 90:586
González-Pérez E, de Jesús YMM, Manuel Ortega-Escobar H et al (2009) Comparative analysis among pathogenic fungal species that cause Gladiolus (Gladiolus grandiflorus Hort) Corm Rot in Mexico. Rev Mex Fitopatol 27:45–52
Fan X, **ao M, Kong F et al (2014) A rare fungal species, Quambalaria cyanescens, isolated from a patient after augmentation mammoplasty—Environmental contaminant or pathogen? PLoS ONE 9:e106949. https://doi.org/10.1371/journal.pone.0106949
Kuan CS, Yew SM, Toh YF et al (2015) Identification and characterization of a rare fungus, Quambalaria cyanescens, isolated from the peritoneal fluid of a patient after nocturnal intermittent peritoneal dialysis. PLoS ONE 10:e0145932. https://doi.org/10.1371/journal.pone.0145932
Delgado-Ramírez CS, Sepúlveda E, Rangel-Montoya EA et al (2023) Heritage grapevines as sources of biological control agents for Botryosphaeria dieback pathogens. Phytopathol Mediterr 62:115–134. https://doi.org/10.36253/phyto-14154
Garcia JF, Lawrence DP, Morales-Cruz A et al (2021) Phylogenomics of plant-associated Botryosphaeriaceae species. Front Microbiol 12:1–18. https://doi.org/10.3389/fmicb.2021.652802
Rangel-Montoya EA, Paolinelli M, Rolshausen PE et al (2021) Characterization of Lasiodiplodia species associated with grapevines in Mexico. Phytopathol Mediterr 60:237–251. https://doi.org/10.36253/phyto-12576
Mateo JJ, Andreu L (2020) Characterization of an exocellular ethanol-tolerant β-glucosidase from Quambalaria cyanescens isolates from unripened grapes. Eur Food Res Technol 246:2349–2357. https://doi.org/10.1007/s00217-020-03578-w
Choi JY, Park AR, Kim YJ et al (2011) Purification and characterization of an extracellular β-glucosidase produced by Phoma sp. KCTC11825BP isolated from rotten mandarin peel. J Microbiol Biotechnol 21:503–508. https://doi.org/10.4014/jmb.1102.02014
Vijaykumar MH, Veeranagouda Y, Neelakanteshwar K, Karegoudar TB (2006) Decolorization of 1:2 metal complex dye Acid blue 193 by a newly isolated fungus, Cladosporium cladosporioides. World J Microbiol Biotechnol 22:157–162. https://doi.org/10.1007/s11274-005-9013-4
Bi R, Lawoko M, Henriksson G (2016) Phoma herbarum, a soil fungus able to grow on natural lignin and synthetic lignin (DHP) as sole carbon source and cause lignin degradation. J Ind Microbiol Biotechnol 43:1175–1182. https://doi.org/10.1007/s10295-016-1783-1
Krishnamoorthy R, Jose PA, Ranjith M et al (2018) Decolourisation and degradation of azo dyes by mixed fungal culture consisted of Dichotomomyces cejpii MRCH 1–2 and Phoma tropica MRCH 1–3. J Environ Chem Eng 6:588–595. https://doi.org/10.1016/j.jece.2017.12.035
Montenecourt BS, Eveleigh DE (1977) Semiquantitative plate assay for determination of cellulase production by Trichoderma viride. Appl Environ Microbiol 33:178–183. https://doi.org/10.1128/aem.33.1.178-183.1977
Acknowledgements
AAM thanks CONACYT for the postdoctoral scholarship funding through “ESTANCIAS POSDOCTORALES POR MÉXICO MODALIDADES 1 Y 2 en la Modalidad: Modalidad 1: Estancia Posdoctoral Académica”, CVU number 385015. Also, AAM thanks to C. Cajica, A. Jiménez-Mu and J. Arriaga-Espinosa for their technical help during the field and laboratory procedures. REAA and RGO thank the company LABCITEC for providing us with their facilities to carry out part of the molecular protocols.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AAM and JRPA designed and conducted the study and wrote the first draft. REAA and RGO performed the molecular approaches. All authors contributed to the manuscript’s final discussion, writing, and revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Argüelles-Moyao, A., Ángeles-Argáiz, R., Garibay-Orijel, R. et al. Isolation and Enzymatic Characterization of Fungal Strains from Grapevines with Grapevine Trunk Diseases Symptoms in Central Mexico. Curr Microbiol 81, 200 (2024). https://doi.org/10.1007/s00284-024-03709-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-024-03709-6