Abstract
Since 2010, porcine epidemic diarrhea virus (PEDV) has received global attention with the emergence of variant strains characterized with high pathogenicity. The pathogen–host interaction after PEDV infection is still unclear. To investigate this issue, high-throughput-based sequencing technology is one of the optimal choices. In this study, we used in vitro transcription sequencing alternative polyadenylation sites (IVT-SAPAS) method, which allowed accurate profiling of gene expression and alternative polyadenylation (APA) sites to profile APA switching genes and differentially expressed genes (DEGs) in IPEC-J2 cells during PEDV variant strain infection. We found 804 APA switching genes, including switching in tandem 3′ UTRs and switching between coding region and 3′ UTR, and 1,677 DEGs in host after PEDV challenge. These genes participated in variety of biological processes such as cellular process, metabolism and immunity reactions. Moreover, 413 genes, most of which are the “focus” genes in interaction networks, were found to be involved in both APA switching genes and DEGs, suggesting these genes were synchronously regulated by different mechanisms. In summary, our results gave a relatively comprehensive insight into dynamic host–pathogen interactions in the regulation of host gene transcripts during PEDV infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine epidemic diarrhea virus (PEDV), an alphacoronavirus, causes acute diarrhea, vomiting, dehydration and weight loss in piglets. Before 2010, PEDV was under control by proper vaccine programs [1, 2]. However, from October 2010, due to the emergence and rapid spreading of the highly pathogenic PEDV variant strains, a large number of suckling piglets in southern China, even in vaccinated herds, presented symptoms such as severe diarrheal disease and higher mortality rates [3]. Moreover, the high morality in infected neonatal piglets leads to enormous economic loss, making PEDV a huge threat to swine industry. From May 2013, a PEDV variant began to emerge and quickly spread throughout the USA and caused numerous deaths in neonatal piglets [4, 2a). After clustering the filtered reads, only 23% of poly(A) sites were mapped to annotated 3′ UTR (Fig. 2b). Based on our dataset, 22% and 6% of poly(A) sites were located in intron and CDS regions, respectively (Fig. 2b). Moreover, 40% and 9% of poly(A) sites were located in intergenic regions and within 1 kb downstream of the beginning of annotated 3′ UTR (Fig. 2b).
IVT-SAPAS reads and poly(A) sites in IPEC-J2 cells. a Genomic location of reads. b Distribution of poly(A) sites. c Genes with different numbers of tandem poly(A) sites. d Boxplot of the median distances between stop codons and poly(A) sites in genes with single poly(A) sites and distances between stop codons and closest poly(A) sites or farthest poly(A) sites in genes with APA in the 3′-end. The elements of box means the upper limit of outliers; 75th percent (Q1); 50th percent (Q2); 25th percent (Q3); the lower limit of outliers. The circles in box plot means outlier. The upper limit of outliers = Q3 + 1.5*(Q3-Q1); the lower limit of outliers = Q1-1.5*(Q3-Q1). (E) PAS usage of poly(A) sites. ucsc: database of University of California, Santa Cruz
We arranged the annotated 3′ UTR APA sites in tandem APA based on the stop codon and combined these data with poly(A) site profiling as previously described [18]. All 10,971 genes were identified to have at least one tandem poly(A) sites, among which 8,080 genes (73.65%) had two or more (Fig. 2c). The average distance between single poly(A) sites and stop codon was about 527 bp (median: 312 bp). The median values of the distance from the stop codon to the proximal or distal poly(A) sites were 259 bp (average: 417 bp) and 1283 bp (average: 1604 bp), respectively (Fig. 2d). The poly(A) signals (PAS) are six-nucleotide cis-acting elements important in 3′ end processing that are located 10–30 nt upstream of the cleavage sites. To date, 12 variants of PAS have been identified, and the canonical types were AAUAAA and its main variant AUUAAA [13]. According to our database, the frequency of canonical PAS was around 46%. About 22% PAS were not identified (Fig. 2e). These results reveal the detailed landscape of poly(A) site usage in IPEC-J2 cells.
Tandem 3′ UTR APA switching in PEDV-infected IPEC-J2 cells
According to different switching sites, APA events can be classified into four general types: tandem 3′ UTR APA and alternative terminal exon APA, which involve cleavage in 3′ UTRs; intronic APA, which is less frequent; and internal exon APA, which is the least frequent type [13]. The tandem 3′ UTR APA is the most frequent APA form. Association of tandem APA with various biological effects has previously been demonstrated, including cancer transformation, embryonic development, immune responses, neuronal activity [12, 14], as well as antiviral innate immune response [15, 16]. Thus, we analyzed the differential use of tandem APA sites located within the 3′ UTR in IPEC-J2 cells infected with PEDV GDS01.
We identified 196, 146, 470, 416, 209, 81 and 75 APA switching genes at 1, 4, 7, 10, 13, 16, 19 hpi, respectively (Fig. 3a), among which 171 genes switched to longer 3′ UTR, 273 genes switched to shorter, while 360 genes showed dynamic switching of 3′ UTR length at different time points, which suggests that these dynamic switching genes may play an active role in the host–pathogen interaction (Fig. 3b). According to the tandem APA switching genes at different time points, we found that 3′ UTR tended to be shortened at the early stage after infection, while the number of genes with lengthened 3′ UTR increased at 7–10 hpi (Fig. 3a), which were consistent with previous research [15]. Then we investigated the biological processes of the tandem 3′ UTR APA switching genes and found that most genes were involved with cellular process, metabolic process and response to stimulus etc., few genes were involved in immune system process (Fig. 3c). After pathway enrichment, the genes with highly significant p values were mainly involved with spliceosome, protein processing in endoplasmic reticulum (ER), Epstein-Barr virus infection, endocytosis pathway etc. (Fig. 3d). For immune response after PEDV infection, several associated pathways were found, such as antigen processing and presentation, TGF-beta signaling pathway, etc. (Fig. 3d).
Tandem APA switching genes in IPEC-J2 cells after PEDV infection. a Summary of tandem APA switching genes between samples at different time points. b Venn diagrams of tandem APA sites switching genes. c Functional classification of tandem APA switching genes. d Pathway analysis of tandem APA switching genes
Dynamic regulation of gene expression in host–pathogen interaction
In addition to measuring tandem 3′ UTR APA variety, SAPAS method could also be used for digital gene expression analysis [19]. We identified 912, 260, 786, 568, 58, 244 and 163 differentially expressed genes at 4, 7, 10, 13, 16, 19 hpi, respectively (Fig. 4a), among which 638 genes were up-regulated, 311 genes were down-regulated and 728 genes were dynamically regulated at different time points (Fig. 4b). These DEGs were clustered into cellular process, metabolic process, response to stimulus and other biological process, few genes were involved in immune system process (Fig. 4c). After the pathway enrichment, metabolic pathways, RNA transport, ribosome, biosynthesis of antibiotics, cell cycle and protein processing in ER pathways were enriched with highly significant p values (Fig. 4d). Furthermore, we found that some noteworthy pathways among these DEGs, such as TGF-beta signaling pathway, RNA transport and degradation, p53 signaling pathway, cell cycle and adherens junction (Fig. 4d).
Endoplasmic reticulum is a major site of protein synthesis, transport and folding [20]. In our analysis, both tandem APA switching genes and differently expressed genes were clustered into protein processing in ER pathway, which suggests that PEDV infection may affect normal function of ER in multiple ways. Ten genes (CANX, DNAJC10, SEC63, UFD1, PLAA, UBE2G2, UBQLN1, BAK1, SSR3, DNAJB11) were selected to detect their dynamic expression with RT-qPCR. The relative expression of related DEGs was confirmed by the RT-qPCR results, which were consistent with the results of SAPAS sequencing (Fig. 5).
RT-qPCR analysis of the dynamic expression of selected genes. IPEC-J2 cells infected with PEDV GDS01 at 0.5 MOI were collected at six time points and RT-qPCRs were performed. The x-axis denotes different time points and the y-axis denotes the relative expression in infected samples comparing to uninfected samples. SAPAs represent the relative expression of genes calculated from IVT-SAPAS sequencing database
APA switching between coding region and 3′ UTR
Besides the 3′ UTR, APA sites are also found in the coding region, thus affecting translation. As described before, if the usage of coding region APA sites is increased, it is referred as CDS-prefer, or UTR-prefer otherwise [16]. A total of 149 genes (FDR < 0.01) showed switching events between coding region and 3′ UTR, and 27 genes showed different tendencies at different time points after infection (Fig. 6a). As the results suggest, genes tend to use more APA sites in 3′ UTR at 1–10 hpi After 10 hpi, the usage of APA sites in coding region prevails (Fig. 6b). APA sites located in the coding region could affect translation and cause premature translation termination. Thus, we explored the function of these CDS-preferred APA genes and found that CDS-preferred genes mainly participate in cellular process and metabolic process (Fig. 6c).
The role of APA and differential expression during PEDV infection
Since both tandem APA switching genes and DEGs showed dynamic regulation during PEDV infection, to explore the relationship between them, we analyzed the two data sets and found that 413 genes (nearly 20%) were subjected to changes of 3′ UTR length and differential expression level during the PEDV infection concurrently (Fig. 7a). These genes are mainly enriched in spliceosome, protein processing in ER and cell cycle pathways (Fig. 7b).
Furthermore, we mapped the gene network connections of tandem APA switching genes and DEGs to explore the potential links between them. We chose cell cycle [21, 22], endoplasmic reticulum protein processing [23], cell junction [24] and PI3K-Akt signaling pathway [25], four pathways that were reported in previous research concerning PEDV infection. As shown in Fig. 8, genes clustered in either alternative polyadenylation or differential expression were mapped to four specific functional networks, while the overlapped genes are preferentially mapped as “focus” genes, such as CDK6, E2F1, DNAJC10, CANX and AKT3.
Discussion
PEDV is one of the pathogens that threatens the global swine industry. Although the pathogenicity of the virus was studied extensively, little is known about the mechanism of the virus–host interaction. Several studies tried to investigate it using proteomics analysis of Vero E6 cells infected with PEDV [8,9,10,11]. Moreover, the global dynamic regulation of PEDV-infected Vero E6 cells at the transcriptional level has been analyzed [7]. Since the regulation of host genes after PEDV infection is a dynamic and complicated process, various types of transcript regulation, including alteration of mRNA quantity and alternative polyadenylation, are involved. In this study, IPEC-J2 cells were used to study the dynamic regulation after PEDV variant strain GDS01 infection. High-throughput-based IVT-SAPAS technology was applied and the differentially expressed genes were profiled.
Extremely complex gene regulatory changes occur in cells after virus infection. On one hand, to replicate in host cells, viruses develop strategies to disturb host gene expression or protein production. On the other hand, cells carry out various defense mechanisms. One way is through affecting sites of 3′ end processing of mRNA, which has consequences for transcript stability and mRNAs translation [13]. As a consequence of alternative polyadenylation, a variety of cellular processes are affected. Since the 3′ UTR serves as a docking platform for posttranscriptional gene regulators, alterations of tandem 3′ UTR APA could influence the stability, localization and translation of the target RNAs. The length of 3′ UTRs influence the translation efficiency, and truncated transcripts lead to truncated proteins. In this work, we just focused on gene expression at the transcript level. In our results, APA switching genes were mainly clustered into cellular processes such as endocytosis, protein processing in ER and spliceosome. Moreover, the same pathways were enriched with the DEGs. Since IPEC-J2 cells were not immune cells, fewer genes affected are associated with immune reaction. Immune pathways such as antigen processing and presentation were found to be promoted in this research.
In this study, APA switching genes and DEGs were found to participate in variety of biological processes such as cellular process, metabolism and immunity reactions, which was consistent with the results of Zhang et al., who analyzed the DEGs of PEDV infection in Vero cells [7]. Several proteome analyses also show that, after PEDV infection, differentially expressed proteins involve in biosynthesis, metabolic and signal transduction processes, such as cell cycle, cell death and survival and virus entry [8,9,10,11]. Immune-related pathways, like TGF-beta signaling pathway, p53 signaling pathway and antigen processing and presentation, were enriched in our results. The expression levels of more immune-related genes and proteins were found to be affected by PEDV infection by Zhang et al. and Sun et al. [7, 10]. In our data, some genes exhibited both APA switching and changes in expression levels, suggesting these genes are regulated by multiple mechanisms during virus infection. Furthermore, the overlapped genes tended to be the “focus” genes in the networks of cellular processes such as cell cycle and protein processing in ER as well as signaling pathways (Fig. 8). These genes may serve as essential genes during PEDV-cell interaction.
The ER serves as a site of protein folding as well as transport [20]. Accumulation of misfolded proteins in the ER during virus infection would cause ER stress. Various research found that a majority of coronaviruses cause ER stress after infection, such as porcine transmissible gastroenteritis virus, severe acute respiratory syndrome virus and infectious bronchitis virus [26,27,28]. For PEDV infection, researchers demonstrated that the viral structural proteins including envelope protein (E) [29], membrane protein (M) [22] and nucleocapsid protein (N) [23] could cause ER stress, resulting in activation of pathways distinct from NF-kB activation. In our gene profiling, we could not find any activation in the NF-kB signaling pathway. Stimulation of immune responses could be studied further in future work.
Cell cycle directs cell proliferation through a series of events. The PEDV structural proteins were shown to be able to prolong the S-phase in a cell cycle [29]. In our study, DEGs and APA switching genes could be mapped to the cell cycle pathway, suggesting the cellular state may serve an important role in PEDV replication. The focus genes exhibiting both APA switching and changes in expression levels may act as more important functional genes. CCNB1, CDK6, ANAPC7 and CDKN2C are main regulators of cell cycle progression, while E2F1 and PCNA participate in DNA replication [30,31,32,33].
Conclusions
Our results reveal a dynamic regulation of gene expression in IPEC-J2 cells after PEDV infection, in which both APA sites switching events and DEGs are involved. About 20% of the significantly changed genes were identified as both APA switching genes and DEGs. These genes tended to act as a “focus genes” in function networks, which may have greater research value in the future. Our results could provide a relatively comprehensive insight into dynamic interaction between host and PEDV virus, which would contribute to future research on PEDV pathogenesis.
Methods
Cells and virus
A porcine small intestinal epithelial cell line IPEC-J2 cells (a kind gift from Prof. Qian Yang, Nan**g Agricultural University, China) were cultured in RPMI 1640 medium (Gibco) containing 10% fetal bovine serum (Gibco) and antibiotics (penicillin 100U/mL and streptomycin 100ug/mL) at 37℃ in 5% CO2. PEDV strain GDS01 (GenBank ID: KM089829.1) [34] was isolated in 2012, Guangdong province, China. The GDS01 was propagated in Vero E6 cells and stored at − 80℃.
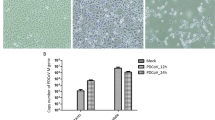
Immunofluorescence analysis (IFA)
IPEC-J2 cells were cultured until 80–90% confluence in 12-well plates and then infected with GDS01 at a multiplicity of infection (MOI) of 0.1. At 7, 13 and 25 h post-inoculation (hpi), cells were collected and washed with PBS before fixation with cold 4% paraformaldehyde for 15 min. The fixed cells were washed twice with PBS and permeabilized with 0.5% Triton X-100 at room temperature (RT) for 15 min. After washing twice with PBS, cells were blocked with 3% (w/v) BSA in PBS at RT for 1 h. Anti-PEDV N protein mouse monoclonal antibody 2B11 [35] (1:1000 diluted in PBST) was used to detect the GDS01-infected cells. After washing thrice with PBST, cells were incubated with FITC-conjugated goat anti-mouse Ig(H + L) (1:500 diluted in PBST) at 37℃ for 1 h. After washing thrice with PBST, fluorescent images were taken under inverted fluorescence microscope (Nikon, Eclipse Ti-U).
Kinetics of PEDV replication in IPEC-J2 cells
Confluent IPEC-J2 cells were washed three times with PBS. After infection with GDS01 at 0.1 MOI for 1 h, the supernatant was replaced by fresh RPMI 1640 medium with 2.5 μg/mL EDTA-free trypsin. At 1, 7, 13, 25 and 37 hpi, the virus was released by freezing and thawing of the cells for three times. The viral titer at each time point was determined by plaque assay. Each experiment was repeated three times independently.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from the GDS01-infected or mock-infected IPEC-J2 cells using TRIzol (Invitrogen) according to the manufacturer’s instruction. cDNA was synthesized using 3 μg total RNA with the Rever Tra Ace qPCR RT Master Mix with gDNA Remover Kit (TOYOBO, Japan) according to the manufacturer’s instruction. qPCR reaction was performed in a final volume of 10 μl with SYBR Premix ExTaq II Kit (Takara) using a Light Cycler 480 real-time PCR system (Roche Diagnostics, Indianapolis, IN, USA). The GAPDH gene was used as a reference gene. Primer sequences in this research are given in Additional file 1.
IVT-SAPAS library preparation and sequencing
Confluent IPEC-J2 cells were washed three times with PBS. After infected with GDS01 at 5 MOI for 1 h, the supernatant was replaced by fresh medium. Mock infection was used as control. Cells were collected at 4, 7, 10, 13, 16, 19 hpi. In total, fourteen samples were used for library preparation. Briefly, total RNA was extracted from samples using TRIzol (Invitrogen). Genomic DNA was removed by using Turbo DNA-free Kit (Ambion) following manufacturer’s instruction. Approximately 2 μg total genomic DNA-free RNA of each sample was randomly fragmented by heating at 94℃ for about 20 min. The first-strand cDNA was synthesized by SuperScript III Reverse Transcriptase Kit (Invitrogen) using an anchored oligo d (T) primer with a 5′ template switching adaptor. Using an anchored oligo dT primer for first-strand synthesis cannot avoid obtaining polyA-rich regions, but this sequence would be filtered out in the reads processing [36]. Then the second-strand cDNA was synthesized by using cDNA synthesis Kit (M-MLV version) (Gubler-Hoffman) according to the manufacturer’s instruction. The cDNA product was purified with Agencourt RNA Clean XP kit (Beckman Coulter). In vitro transcription was done by using RiboMAX™ Large Scale RNA Production System-T7 (Promega). After removing template cDNA with RQ1 RNase-Free DNase (Promega), the RNA product was purified with Agencourt RNA Clean XP kit (Beckman Coulter). Illumina adaptor B with 6-mer barcode and modified oligo d(T) tagged illumine adaptor A were used for PCR amplification following the instruction of High Fidelity Platinum Taq DNA Polymerase Kit (Invitrogen). Then fragments of 250–500 bp in size of the PCR products were selected and purified by Agencourt Ampure magnetic bead (Beckman Coulter) according to the manufacturer’s instruction. The average size was determined by Agilent 2100 bioanalyzer (Agilent Genomics). Then the final pooled fragments were quantified and sequenced from the 3′ end with Hiseq 2500 (Illumina). The details of primers were shown in Additional file 1.
Data analysis
Data from the SAPAS-libraries was analyzed as described before [18]. Briefly, generated raw reads were trimmed, filtered, and mapped to the sus scrofa genome (http://hgdownload.soe.ucsc.edu/goldenPath/susScr3/bigZips/). After internal priming filtering [36], the resulting uniquely mapped reads were clustered to define poly(A) sites.
Two types of APA switching events, named tandem APA switching and switching between coding regions and 3′ UTR, were considered in this study. To test tandem APA switching, poly(A) site usage between infected and uninfected cells were identified by the linear trend test and the independence test. As to test switching between coding regions and 3′ UTR, APA sites within a gene were classified into two groups according to their locations, referring as coding region and 3′ UTR. Then Chi-squared test was introduced to evaluate whether the switching that occurred within these two regions is significant. If the usage of the APA sites in the coding region is increased, it is determined as CDS-prefer, otherwise is UTR-prefer [16].
To analyze gene expression level, the ratio between infected and uninfected samples was compared with chi-squared test. If gene with significant p value corresponded to a false discovery rate < 0.05 and fold change > 2, they were identified as significant DEGs.
KEGG pathway information was extracted from KEGG MAPPER (http://www.genome.jp/kegg/tool/map_pathway1.html) on the KEGG website.
The resulting p values were corrected in the Benjamini–Hochberg sense. Bubble charts of pathways were drawn with R package ggplot2. The protein interaction was analyzed with SRING and then drawn with Cytoscape.
Abbreviations
- PEDV:
-
Porcine epidemic diarrhea virus
- IPEC-J2:
-
Porcine small intestinal epithelial cell line
- hpi:
-
Hours post-inoculation
- IVT-SAPAS:
-
In vitro Transcription sequencing alternative polyadenylation sites
- APA:
-
Alternative polyadenylation
- DEGs:
-
Differentially expressed genes
- PASs:
-
Poly(A) signals
- DEPs:
-
Differentially expressed proteins
- CPEs:
-
Cytopathic effects
- MOI:
-
Multiplicity of infection
- ucsc:
-
Database of University of California, Santa Cruz
- ER:
-
Endoplasmic reticulum
- RT:
-
Room temperature
- RT-qPCR:
-
Quantitative real-time PCR
References
Kusanagi K, Kuwahara H, Katoh T, Nunoya T, Ishikawa Y, Samejima T, Tajima M (1992) Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci 54(2):313–318. https://doi.org/10.1292/jvms.54.313
Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q (2012) New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis 18(8):1350–1353. https://doi.org/10.3201/eid1808.120002
Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX (2012) Outbreak of porcine epidemic diarrhea in suckling piglets. China Emerg Infect Dis 18(1):161–163. https://doi.org/10.3201/eid1801.111259
Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, and Meng XJ (2013) Origin, evolution, and genoty** of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4(5):e00737–13. DOI: https://doi.org/10.1128/mBio.00737-13.
Tian PF, ** YL, **ng G, Qv LL, Huang YW, Zhou JY (2014) Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis 20(10):1735–1738. https://doi.org/10.3201/eid2010.140338
Wang D, Fang L, **ao S (2016) Porcine epidemic diarrhea in China. Virus Res 226:7–13. https://doi.org/10.1016/j.virusres.2016.05.026
Zhang H, Liu Q, Su W, Wang J, Sun Y, Zhang J, Shang K, Chen Z, Cheng S, Wu H (2018) Genome-wide analysis of differentially expressed genes and the modulation of PEDV infection in Vero E6 cells. Microb Pathog 117:247–254. https://doi.org/10.1016/j.micpath.2018.02.004
Guo X, Hu H, Chen F, Li Z, Ye S, Cheng S, Zhang M, He Q (2016) iTRAQ-based comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of porcine epidemic diarrhea virus. J Proteomics 130:65–75. https://doi.org/10.1016/j.jprot.2015.09.002
Lin H, Li B, Chen L, Ma Z, He K, Fan H (2017) Differential Protein Analysis of IPEC-J2 Cells Infected with Porcine Epidemic Diarrhea Virus Pandemic and Classical Strains Elucidates the Pathogenesis of Infection. J Proteome Res 16(6):2113–2120. https://doi.org/10.1021/acs.jproteome.6b00957
Sun D, Shi H, Guo D, Chen J, Shi D, Zhu Q, Zhang X, Feng L (2015) Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J Virol Methods 218:27–39. https://doi.org/10.1016/j.jviromet.2015.03.002
Zeng S, Zhang H, Ding Z, Luo R, An K, Liu L, Bi J, Chen H, **ao S, Fang L (2015) Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics 15(11):1819–1828. https://doi.org/10.1002/pmic.201400458
Batra R, Manchanda M, Swanson MS (2015) Global insights into alternative polyadenylation regulation. RNA Biol 12(6):597–602. https://doi.org/10.1080/15476286.2015.1040974
Elkon R, Ugalde AP, Agami R (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14(7):496–506. https://doi.org/10.1038/nrg3482
Huang G, Huang S, Wang R, Yan X, Li Y, Feng Y, Wang S, Yang X, Chen L, Li J, You L, Chen S, Luo G, Xu A (2016) Dynamic Regulation of Tandem 3’ Untranslated Regions in Zebrafish Spleen Cells during Immune Response. J Immunol 196(2):715–725. https://doi.org/10.4049/jimmunol.1500847
Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P, Chen S, Xu A (2017) The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun 8:14605. https://doi.org/10.1038/ncomms14605
Li JHL, Zhang Y, Xue CY, Cao YC (2017) A novel method for genome-wide profiling of dynamic host-pathogen interactions using 3’ end enriched RNA-seq. Scientific Reports 7(1):8681. https://doi.org/10.1038/s41598-017-08700-9
Wei Y, Li J, Zhang Y, Xue C, Cao Y (2018) Tandem 3’ UTR Patterns and Gene Expression Profiles of Marc-145 Cells During PRRSV Infection. Virol Sin 33(4):335–344. https://doi.org/10.1007/s12250-018-0045-y
Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A (2011) Differential genome-wide profiling of tandem 3’ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21(5):741–747. https://doi.org/10.1101/gr.115295.110
Fu YG, Sun Y, Li YX, Li J, Rao XQ, Chen C, Xu AL (2011) Differential genome-wide profiling of tandem 3 ’ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21(5):741–747
Schwarz DS, Blower MD (2016) The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73(1):79–94. https://doi.org/10.1007/s00018-015-2052-6
Ye S, Li Z, Chen F, Li W, Guo X, Hu H, He Q (2015) Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes 51(3):385–392. https://doi.org/10.1007/s11262-015-1257-y
Xu XG, Zhang HL, Zhang Q, Dong J, Huang Y, Tong DW (2015) Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol 59(3):265–275. https://doi.org/10.4149/av_2015_03_265
Xu X, Zhang H, Zhang Q, Huang Y, Dong J, Liang Y, Liu HJ, Tong D (2013) Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol 164(3–4):212–221. https://doi.org/10.1016/j.vetmic.2013.01.034
Luo X, Guo L, Zhang J, Xu Y, Gu W, Feng L, and Wang Y (2017) Tight Junction Protein Occludin Is a Porcine Epidemic Diarrhea Virus Entry Factor. J Virol 91(10). DOI: https://doi.org/10.1128/JVI.00202-17.
Kong N, Wu Y, Meng Q, Wang Z, Zuo Y, Pan X, Tong W, Zheng H, Li G, Yang S, Yu H, Zhou EM, Shan T, Tong G (2016) Suppression of Virulent Porcine Epidemic Diarrhea Virus Proliferation by the PI3K/Akt/GSK-3alpha/beta Pathway. PLoS ONE 11(8):e0161508. https://doi.org/10.1371/journal.pone.0161508
Fung TS, Liao Y, and Liu DX (2016) Regulation of Stress Responses and Translational Control by Coronavirus. Viruses 8(7). DOI: https://doi.org/10.3390/v8070184.
Fung TS, Huang M, Liu DX (2014) Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res 194:110–123. https://doi.org/10.1016/j.virusres.2014.09.016
Cossart P and Helenius A (2014) Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol 6(8). DOI: https://doi.org/10.1101/cshperspect.a016972.
Xu X, Zhang H, Zhang Q, Dong J, Liang Y, Huang Y, Liu HJ, Tong D (2013) Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol J 10:26. https://doi.org/10.1186/1743-422X-10-26
Gao H, Yin Y, Qian A, Guo R, Qi J (2019) LncRNA LINC00974 Upregulates CDK6 to Promote Cell Cycle Progression in Gastric Carcinoma. Cancer Biother Radiopharm 34(10):666–670. https://doi.org/10.1089/cbr.2019.2904
Choi EH, Kim KP (2019) E2F1 facilitates DNA break repair by localizing to break sites and enhancing the expression of homologous recombination factors. Exp Mol Med 51(9):1–12. https://doi.org/10.1038/s12276-019-0307-2
Chai N, **e HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, Nie YZ, Wu KC, Yang AG, Zhang R (2018) FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun 500(4):924–929. https://doi.org/10.1016/j.bbrc.2018.04.201
Boehm EM, Gildenberg MS, Washington MT (2016) The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 39:231–254. https://doi.org/10.1016/bs.enz.2016.03.003
Hao J, Xue C, He L, Wang Y, Cao Y (2014) Bioinformatics insight into the spike glycoprotein gene of field porcine epidemic diarrhea strains during 2011–2013 in Guangdong. China Virus Genes 49(1):58–67. https://doi.org/10.1007/s11262-014-1055-y
Gong L, Lin Y, Qin J, Li Q, Xue C, Cao Y (2018) Neutralizing antibodies against porcine epidemic diarrhea virus block virus attachment and internalization. Virol J 15(1):133. https://doi.org/10.1186/s12985-018-1042-3
Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, Chen L, Chen S, Xu A (2012) Dynamic landscape of tandem 3’ UTRs during zebrafish development. Genome Res 22(10):1899–1906. https://doi.org/10.1101/gr.128488.111
Acknowledgements
We would like to thank Prof. Qian Yang (Nan**g Agricultural University, China) for the IPEC-J2 cell line.
Funding
This work was supported by National Key Research and Development Program (2016YFD0500101).
Author information
Authors and Affiliations
Contributions
YC, YZ, XW, LG and JL designed the study. XW performed the experiments. JL and XW analyzed the data; XW prepared the figures and tables; XW and YZ wrote the main manuscript. YC and YZ checked and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Edited by Simon D. Scott.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Additional files 1: Primers for RT-qPCR and PCR amplification at the last step in preparation IVT-SAPAS library.
Rights and permissions
About this article
Cite this article
Wei, X., Li, J., Zhang, Y. et al. Profiling of alternative polyadenylation and gene expression in PEDV-infected IPEC-J2 cells. Virus Genes 57, 181–193 (2021). https://doi.org/10.1007/s11262-020-01817-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-020-01817-6