Abstract
Avipoxviruses are thought to be restricted to avian hosts and considered significant pathogens that may impact the conservation of many birds. However, reports of avipoxvirus-like viruses from reptiles suggest that cross-species transmission, within birds and other species, may be possible. The vast majority of avipoxviruses in wild birds remain uncharacterised and their genetic variability is unclear. Here, cutaneous pox lesions were used to recover a novel full-length crowpox virus genome from an Australian little crow (Corvus bennetti), followed by the detection of immature and intracellular mature virions using electron microscopy. The CRPV genome was 328,768 bp in length and contained 403 predicted open-reading frames. While 356 of the ORFs of CRPV genome had the greatest similarity with other avipoxviruses gene products, a further 47 ORFs were novel. Subsequent phylogenetic analyses showed that the CRPV was most closely related to other avipoxviruses isolated from passerine and marine bird species and demonstrated the highest sequence similarity with an albatrosspox virus (84.4%). Considering the sequence similarity observed between CRPV and other avipoxviruses and phylogenetic position, this study concluded that the CRPV to be a distinct available candidate of avipoxviruses.
Similar content being viewed by others
Introduction
Avipoxviruses are large, double-stranded DNA (dsDNA) viruses belonging to the genus Avipoxvirus (family Poxviridae, subfamily Chordopoxvirinae) that may cause proliferative, diphtheritic or systemic lesions in birds1,2. Avipoxviruses represent a diverse virus group, that may infect most avian species. Evidence of poxvirus infection has been found in at least 329 avian species and 20 orders of wild and domestic bird species3,4, with many more avian hosts considered susceptible. In general, avipoxviruses appear to have been present in bird populations for continuous periods, leading to low levels of infection and relatively mild disease. However, where poxviruses have been introduced to naïve bird populations, they have the potential to cause explosive outbreaks of severe disease with high morbidity and mortality, as occurred in Hawaii, the Galapagos and the Canary Islands5,6.
According to the International Committee on Taxonomy of Viruses (ICTV)7 there are currently 12 species approved under the genus Avipoxvirus: Canarypox virus, Flamingopox virus, Fowlpox virus, Juncopox virus, Mynahpox virus, Penguinpox virus, Pigeonpox virus, Psittacinepox virus, Quailpox virus, Sparrowpox virus, Starlingpox virus and Turkeypox virus. A further two viruses, crowpox virus and peacockpox virus, are putative members of the genus Avipoxvirus, but have not yet been approved as species by the ICTV. There are currently a limited number of complete avipoxvirus genome sequences from ICTV-recognised species available in GenBank. These include a canarypox virus (CNPV)8, a South African strain of pigeonpox virus (FeP2)9, a penguinpox virus (PEPV)9, a Hungarian strain of turkeypox virus (TKPV)10, an American strain of fowlpox virus (FWPV)11, a European strain of fowlpox virus (FP9)12, and an additional eight complete genome sequences of FWPV that have been published since 201813,14,15. There are also six further complete avipoxvirus genomes: two shearwaterpox viruses (SWPV1 and SWPV2)16, two magpiepox viruses (MPPV and MPPV2)17,18, a mudlarkpox virus (MLPV)19, penguinpox virus 2 (PEPV2)20, two albatrosspox virus (ALPV and ALPV2)21,22, and a poxvirus in house finches (Haemorhous mexicanus)23 available in GenBank that are not yet ICTV recognised species.
Avipoxvirus infection in the family Corvidae has been identified in an adult American crow (Corvus brachyrhynchos) in the United States of America24,25. Several recent studies have also characterised poxvirus infections in the cutaneous lesions of Australian passerine bird species including the Australian magpie (Gymnorhina tibicen)17,18, and the mudlark (Grallina cyanoleuca)19. However, there are no sequence data of crowpox virus from the Australian little crow (Corvus bennetti), and consequently, its genetic and phylogenetic relationships with other avipoxviruses are not well known. The aim of this study was to identify and characterise the genome sequence of crowpox virus (CRPV) from an Australian little crow sourced from Victoria in 2021.
Results
Genome of crowpox virus (CRPV)
We determined the complete genome sequence of CRPV as a linear double-stranded DNA molecule of 328,768 bp in length (GenBank accession no. ON408417). The CRPV genome contained a large central coding region surrounded by two matching inverted terminal repeat (ITR) regions, constituting 4052 bp each (coordinates 1-4052 sense and 324,717–328,768 antisense orientation) like other characterised avipoxviruses11,16,17,19,26. Each of the inverted repeats constituted arrays of direct repeats, and six tandem repeats were detected within each inverted terminal repeat region. These consisted of a 103 bp, 85 bp, 60 bp, 9 bp and two 42 bp repeat unit and sharing approximately 97–100% nucleotide identity. The CRPV genome showed the highest nucleotide identity (84.4%) with the pathogenic avipoxvirus ALPV, isolated from an endangered northern royal albatross (Diomedea sanfordi) in 1997 (GenBank accession no. MW365933)22 (Table 1), followed by PEPV2 (84.3%), SWPV2 (83.4), CNPV (83.3%), and MPPV2 (82.6%). The A + T content of the CRPV genome was 71.3%, which was comparable to other sequenced avipoxviruses (Table 1).
Genome annotation and comparative analyses of CRPV
The CRPV genome predicted to enclose 403 open reading frames (ORFs) encoding proteins ranging from 30 to 1945 amino acids in length, that were numbered from left to right (Fig. 1 and Supplementary Table S1). Among them, five predicted ORFs were found within the ITRs and were thus present as diploid copies. Comparative analysis of the predicted ORF sequences showed that 356 had the greatest similarity with other ChPV gene products (E value ≤ 10−5) (Fig. 1 and Supplementary Table S1). Among these predicted genes, 166 genes showed the highest similarity to a CNPV8, followed by 69 genes to MPPV17, 48 genes to SWPV216, 32 genes to a recently sequenced FIPV23 and 19 genes to PEPV220. A further seven (ORF-114, -117, -284, -363, -387, -397 and -398) showed highest similarity to MLPV, five (ORF-103, -113, -215, -219 and -279) to ALPV, five (ORF-108, -132, -210, -282 and -290) to ChePV1, two (ORF-128 and -129) to FWPV, one (ORF-329) to Hawaiian goose poxvirus (HGPV196), one (ORF-130) to SWPV1, and one (ORF-023) to FGPV (Fig. 1 and Supplementary Table S1).
Genomic illustration of CRPV in comparison with CNPV was visualised using Geneious Prime® (version 2022.1.1). The arrows depict the direction of transcription of genes and open reading frames (ORFs). Each ORF of CRPV genome is colour coded based on homology to other avipoxviruses, as indicated by the key in the legend.
Remarkably, the CRPV genome contained 47 predicted protein-coding genes that were unique based on the NR protein database using BLASTX and BLASTP27. These unique ORFs encoded proteins of 30 to 100 amino acids in length (Fig. 1 and Supplementary Table S1). Among them, 18 unique CRPV protein-coding ORFs (ORF-006, -007, -016, -017, -050, -060, -076, -099, -100, -124, -127, -138, -196, -276, -280, -283, -295, -366) were predicted to contain a single transmembrane helix, and software packages predicted that ORF-052 contained three transmembrane helices (Supplementary Table S1). We did not find any significant homology with known proteins for the unique ORFs encoded in the CRPV genome when using Phyre2, HHpred or SWISS-MODEL, which may result from the lack of closely related structures in these databases.
Dot plot analyses were used to compare the CRPV genome with other selected avipoxviruses. The CRPV genome was highly syntenic with ALPV, CNPV, SWPV2, FIPV and PEPV2 (Fig. 2A–E); however, a difference in synteny was observed (Fig. 2A–E, highlighted as black arrows), mainly due to the absence of two large additional copies of variola B22R gene family proteins and a hypothetical protein coding gene covering approximately 16kbp. However, the CRPV genome demonstrated significant differences in the entire genome compared to other complete avipoxviruses including FeP2 and TKPV (Fig. 2F–G).
Dot plots of the CRPV genome (x-axis) versus other poxvirus genomes (y-axis). (A) CRPV versus ALPV, (B) CRPV versus CNPV, (C) CRPV versus SWPV2, (D) CRPV versus FIPV, (E) CRPV versus PEPV2, (F) CRPV versus FeP2 and (G) CRPV versus TKPV (refer to Table 3 for virus details and GenBank accession numbers). The Classic colour scheme was chosen in Geneious (version 22.1.1) for the dot plot lines according to the length of the match, from blue for short matches to red for matches over 100 bp long. Window size = 12.
Core/conserved ORFs
Similar to other chordopoxviruses (ChPVs) the CRPV genome contained 89 conserved core genes, which are involved in essential functions such as replication, transcription and virion assembly (Supplementary Table S1; highlighted with bold font). The number of conserved ChPV genes is considered to range between 83 and 903,9,28,29, which is consistent with the findings in the CRPV genome. Among them, nine of the predicted ORFs (CRPV-143, -166, -193, -211 -244, -307, -317, -319 and -327) were truncated mostly with a single residue compared to CNPV, which may warrant further studies to determine whether they are expressed and functional. Based on a recent study by Carulei et al.3, we also searched for a further 47 genes that are conserved in avipoxviruses (Table 2). We predicted the CRPV genome would also contain these 47 conserved ORFs (Table 2), and eight of the genes (CRPV-049, -125, -131, -167, -173, -323, -324 and -341) were found to be truncated compared to a closely related canarypox virus (CNPV).
Multigene families
Avipoxviruses are the largest ChPVs and contain several, large, multigene families with immune related functions comprising up to 50% of the genome3,9. The copy numbers of each of the 14 multigene families identified in the CRPV genome compared with the other selected sequenced avian poxvirus genomes, including the recently characterised genomes of ALPV2, ALPV, MPPV2 and PEPV2 (Supplementary Table S2). CRPV has a relatively higher number of multigene families (156 gene copies) compared to the closely related avipoxviruses such as ALPV, CNPV and SWPV2 (total of 139, 137 and 124 gene copies, respectively). The copy number of ankyrin repeat, B22R, C4L/C10L, CC chemokine and TGF-β family genes were relatively higher in the CRPV genome compared to CNPV. However, the copy number of N1R/p28 and C-type lectin genes were significantly lower in the CRPV genome compared to CNPV.
Evolutionary relationships of CRPV
Phylogenetic reconstruction using concatenated amino acid sequences of selected conserved ChPV genes provides clear evidence for the inclusion of CRPV in the genus Avipoxvirus. In the maximum likelihood (ML) tree (Fig. 3), CRPV was located within a subclade B1 encompassing avipoxviruses isolated from several passerine bird species, a wedge-tailed shearwater (A. pacificus), northern royal albatross (Diomedea sanfordi), and yellow-eyed penguin (Megadyptes antipodes) with 100% bootstrap support. In the sub-clade B1, FIPV is basal to known avipoxviruses, suggesting that all the avipoxviruses under this subclade evolved from the ancestral house finches (Haemorhous mexicanus) from where MLPV followed by CRPV viruses evolved, and later to the other avian hosts. Using the same set of concatenated protein sequences, we found that the maximum inter-lineage sequence identity values ranged from 98.7 to 97.9% among avipoxviruses under subclade B1, which reflected the phylogenetic position of this novel avipoxvirus sequenced from an Australian little crow, and further inferred that these viruses likely originated from a common ancestor. Furthermore, it was also evidenced that there were many avipoxviruses evolutionarily linked with crowpox virus sequenced in this study when we compared using partial nucleotide sequences of the DNA polymerase gene (Supplementary Fig. S1) and p4b gene (Supplementary Fig. S2). Among them, avipoxviruses isolated from an American crow (Corvus brachyrhynchos) and a canary (Serinus canaria) in Chile25, Australian magpies17,18, a gray-crowned rosy finch (Leucosticte tephrocotis) and a Swainson's thrush (Catharus ustulatus) in the USA25 were the closest evolutionary link with the CRPV isolated in this study.
Phylogenetic relationships between CRPV and other chordopoxviruses. A maximum likelihood (ML) tree was constructed from multiple alignments of the concatenated amino acid sequences of the selected nine poxvirus core proteins using CLC Genomics Workbench (version 9.0.1). The numbers on the left show bootstrap values as percentages. The labels at branch tips refer to virus species, followed by GenBank accession numbers and abbreviated species names in parentheses. The position of CRPV is highlighted using pink text. Details of the poxviruses used in the phylogenetic tree are in Table 3. Saltwater crocodile poxvirus 1 (SwCRV1; MG450915)30 was used as an outgroup. Major clades and sub-clades are designated according to Gyuranecz et al. (2013)25.
Evidence of poxvirus particles in cutaneous pox tissue
Using transmission electron microscopy (TEM) analysis, crowpox virus particles were identified in the sample sourced from the Australian little crow. It showed the presence of intracellular mature virion (IMV) that was brick-ovoid shaped with regular spaced thread-like ridges comprising the exposed surface and was approximately 250 × 230 nm (Fig. 4A), and immature virion was brick shaped, measuring approximately 135 × 125 nm in diameter (Fig. 4B).
Discussion
This paper documents a novel avipoxvirus, crowpox virus (CRPV), with the complete genome sequence obtained directly from naturally occurring pox lesions in an Australian little crow. Applying various approaches for genomic comparison and transmission electron microscopic analysis, the CRPV genome was shown to be architecturally consistent with other avipoxviruses in terms of genome size, AT content and predicted ORFs; however, it was distinct from other avipoxvirus genomes in multiple ways. Overall, the DNA sequence of CRPV was significantly different from other avipoxviruses but had the closest similarity with the pathogenic avipoxvirus, albatrosspox virus (ALPV), followed by penguinpox virus 2 (PEPV2; 84.3%), shearwaterpox virus 2 (SWPV2; 83.4), canarypox virus (CNPV; 83.3%), and magpiepox virus 2 (MPPV2; 82.6%). The novel CRPV genome contained 47 predicted genes that are not found in any other poxvirus, as well as several ORFs that were so truncated/fragmented as to probably cause them to be non-functional. Overall, the CRPV was sufficiently genetically different to other previously classified avipoxviruses to be considered as a distinct new virus species under the genus Avipoxvirus.
Phylogenetic tree analysis demonstrated that the subclade B1 consisting of avipoxviruses isolated from several passerine bird species including this Australian little crow, as well as seabirds including a wedge-tailed shearwater, a northern royal albatross, and a yellow-eyed penguin, supports evidence that this CRPV most likely originated from avipoxvirus isolated from other birds in the order Passeriformes (Fig. 3). The basal position of a recently sequenced avipoxvirus from house finches in the USA (subclade B1, Fig. 3), from where mudlarkpox virus (MLPV) followed by CRPV viruses have likely evolved provides evidence that the evolution of avipoxviruses in Passeriformes is not well understood, given the divergent geographical distribution of the host species. It is possible that Australian passerine birds may be host to many as yet undiscovered avipoxviruses.
Much is still unknown about the host spectrum and epidemiology of poxviruses, in particular for Australian avifauna. Although there is some evidence that poxviruses infecting Australian wild birds including magpies are transmissible to other magpies, but not to chickens, turkeys, pigeons, or canaries after experimental inoculation31,32, their mode of transmission in Australian avifauna is not well understood. Previous studies suggest that avipoxviruses can be transmitted between birds in several ways: (1) via direct contact with infected birds through broken skin; (2) through contact between skin breaches and contaminated objects including perches; (3) by aerosol transmission4,33, and (4) via haematophagous arthropods including mosquitoes, which are efficient mechanical vectors through contaminated mouthparts34,35. These factors may indicate a potential scenario for crowpox virus transmission that merits further attention. At an individual level, poxvirus infections in wild birds can cause primary disease that may be severe in some cases, leading to decreased foraging and mobility. In affected birds, avipoxvirus infection can lead to two different forms of disease. The most common disease is characterised by a proliferative ‘wart-like’ lesions that are commonly restricted to the eyes, beak or unfeathered skin of the body (so-called ‘dry’ pox), in which secondary bacterial and fungal infections may aggravate the birds' condition. The second form of poxviral infection is the ‘wet’ or ‘diphtheritic’ form, characterised by lesions on the mucous membranes of the upper alimentary and respiratory tracts2,36. Avipoxvirus infection in bird can also reduce ability to care for young, and affect vision and/or feeding ability, making them prone to predation, and significantly affecting welfare2,36,37, but in some cases, birds are likely sub clinically infected. The repeated occurrence of avian family or order-specific grou** within certain avipoxvirus clades indicates a marked role of host adaptation, while the sharing of poxvirus species within prey-predator systems (e.g., pigeonpox in raptors)25 indicates the potential for cross-species infection and limited host adaptation25. At a population level, these may have serious implications, especially for endangered or endemic species, and hence further studies into the evolution of avipoxviruses in non-model hosts warrants further investigation.
Conclusions
The novel complete genome sequence of CRPV reported here has enhanced the genomic information for the Avipoxvirus genus, contributing to our understanding of the avipoxviruses more generally, as well as tracking poxvirus evolution in a non-model avian species. By assessing the sequence similarity between CRPV and other avipoxviruses, we concluded that the CRPV complete genome described should be considered a separate avipoxvirus species. Additional investigations will be required to better understand relevant host–pathogen dynamics including routes of transmission and factors leading to infection, associated pathology, and disease prevalence.
Methods
Sampling, ethical consideration and extraction of DNA
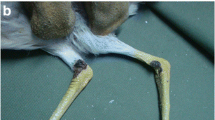
Samples were obtained from a little crow (Corvus bennetti) that was euthanised by inhalational general anaesthesia with isoflurane (IsoFlo, Zoetis Australia Pty Ltd) in oxygen followed by intravenous injection of pentabarbitone sodium (Lethabarb Euthanasia Injection, Virbac Australia Pty Ltd) due to untreatable septic arthritis secondary to proliferative pox lesions by a registered veterinarian at The Unusual Pet Vets, Frankston, Victoria. Animal sampling (ID: 122740) was carried out by the attending veterinarian for the investigation of crusty pox lesions affecting the legs and eyes. Collected samples were stored at − 20 °C until further processing. For DNA extraction, the crusty pox lesion material was aseptically dissected and mechanically homogenised in lysis buffer using disposable tissue grinder pestles and transferred into a 1.5 mL microcentrifuge tube (Eppendorf). Total genomic DNA was isolated according to the established methods38,39 using a ReliaPrep gDNA Tissue Miniprep System (Promega, USA).
Library construction and sequencing
A total of 250 ng of extracted genomic DNA was used to prepare the library using the protocol adapted previously using the Illumina DNA Prep (Illumina, San Diego, CA, USA)40. The quality and quantity of the prepared library was assessed using an Agilent Tape Station (Agilent Technologies) by the Genomic Platform, La Trobe University. The prepared library was sequenced with the sequencing reads length of 150-bp paired-end on Illumina® NovaSeq platform according to the manufacturer's instructions through the Australian Genome Research Facility, Melbourne.
Genome assembly and annotation
The resulting 39.8 million raw sequence reads were used to assemble the complete genome of CRPV, using CLC Genomics Workbench (version 9.0.1, CLC bio, a QIAGEN Company, Prismet, Aarhus C, Denmark) and Geneious Prime® (version 2022.1.1, Biomatters, New Zealand), as described previously16,17,20,26,41. Briefly, the sequences were processed to remove Illumina adapters, low quality reads and ambiguous bases. Trimmed sequence reads were mapped against the chicken genome (Gallus gallus, GenBank accession number NC_006088.5) to remove potential host DNA contamination. In addition, reads were further mapped to the Escherichia coli bacterial genomic sequence (GenBank accession no. U00096) to remove possible bacterial contamination. A total of 30.2 million cleaned and unmapped reads were used as input data for de novo assembly using CLC Genomics Workbench (version 9.0.1). This resulted in the generation of a 328,768 bp genome with an average coverage of 1182.51x. The genome was annotated according to the previously published protocol using Geneious software (version 2022.1.1). Open reading frames (ORFs) longer than 30 amino acids, with a methionine start codon (ATG) and minimal overlap with other ORFs (not exceeding 50% of one of the genes), were selected and annotated. ORFs shorter than 30 amino acids that had been previously annotated in other poxvirus genomes were also included. Similarity BLAST searches were performed on the predicted ORFs and were annotated as potential genes if predicted ORFs showed significant sequence similarity to known viral or cellular genes (BLAST E value ≤ e−5)27. The direct tandem repeats were detected using the Tandem Repeats Finders42.
To predict the function of putative unique ORFs identified in this study, the derived protein sequence of each ORF was searched using multiple applications to identify conserved domains or motifs. Transmembrane helices were searched using the TMHMM package (version 2.0)43 and TMpred44. Additionally, searches for conserved secondary structure (HHpred)45 and protein homologs were conducted using Phyre246 and SWISS-MODEL47.
Comparative genomics
Genomic features of the newly sequenced CRPV were visualised using Geneious Prime® (version 2022.1.1). Sequence similarity percentages between CRPV and representative ChPV complete genome sequences were determined using tools available in Geneious (version 2022.1.1). Dot plots were created based on the EMBOSS dottup program in Geneious software, with word size = 1248.
Phylogenetic analyses
Phylogenetic analyses were performed using the CRPV genome sequence determined in this study, together with other selected ChPV genome sequences available in GenBank (Table 3). The amino acid sequences of nine poxvirus core proteins (RNA polymerase subunit RPO132, RNA polymerase subunit RPO147, mRNA cap** enzyme large subunit, RNA polymerase-associated protein RAP94, virion core protein P4a, virion core protein P4b, early transcription factor large subunit VETFL, NTPase, and DNA polymerase) were concatenated and aligned using MAFTT (version 7.450) with the G-INS-i (gap open penalty 1.53; offset value 0.123) algorithm implemented in Geneious Prime® (version 2022.1.1, Biomatters, New Zealand). Nucleotide sequences of the partial DNA polymerase and partial p4b genes, as well as concatenated amino acid sequences of the selected nine poxvirus core proteins, were aligned as described previously19 using the MAFTT L-INS-I algorithm implemented in Geneious Prime® (version 2022.1.1) (version 7.388)49. To determine the best-fit model to construct phylogenetic analyses, a model test was performed using CLC Genomics Workbench (version 9.0.1), which favoured a general-time-reversible model with gamma distribution rate variation and a proportion of invariable sites (GTR + G + I). Phylogenetic analyses for nucleotide sequences were performed under the GTR substitution model, but the WAG substitution model was chosen for concatenated amino acid sequences with 1000 bootstrap replicates in CLC Genomic Workbench (version 9.0.1).
Transmission electron microscopy
Cutaneous pox lesions were suspended 1:10 in phosphate-buffered saline (PBS), homogenised, clarified and adsorbed onto 400-mesh copper EM grids, before staining and imaging on a JEOL JEM-2100 transmission electron microscope as previously described26,30.
Institutional review board statement
The animal handling and procedures were conducted in accordance with ARRIVE guidelines for experimental procedures. Briefly, samples were obtained from a little crow (Corvus bennetti) that was euthanatised due to untreatable septic arthritis secondary to proliferative pox lesions by a registered veterinarian at The Unusual Pet Vets, Frankston, Victoria. The animal was necropsied by the same registered veterinarian for routine diagnostic purposes. All other methods were performed in accordance with the standard guidelines and regulations for PC2 laboratory. The Animal Ethics Committee at La Trobe University was informed that findings from the diagnostic material were to be used in a publication, and a formal waiver of ethics approval was granted.
Data availability
The complete genome sequence and associated datasets generated during this study were deposited in GenBank under the accession number ON408417. Raw sequencing data from this study has been deposited in the NCBI Sequence Read Achieve (SRA) under the accession number SRR19117728 (BioProject ID: PRJNA835616, BioSample accessions: SAMN28105687) (http://www.ncbi.nlm.nih.gov/sra/).
References
Tripathy, D. N. & Reed, W. M. in Diseases of Poultry 333–349 (2013).
Niemeyer, C. et al. Two different avipoxviruses associated with pox disease in Magellanic penguins (Spheniscus magellanicus) along the Brazilian coast. Avian Pathol. 42, 546–551. https://doi.org/10.1080/03079457.2013.849794 (2013).
Carulei, O., Douglass, N. & Williamson, A.-L. Comparative analysis of avian poxvirus genomes, including a novel poxvirus from lesser flamingos (Phoenicopterus minor), highlights the lack of conservation of the central region. BMC Genom. 18, 947. https://doi.org/10.1186/s12864-017-4315-0 (2017).
van Riper, C. & Forrester, D. J. Avian pox. In Infectious Diseases of Wild Birds (eds Thomas, N. J. et al.) (Wiley Blackwell Publishing, Oxford, 2007).
Young, L. C. & VanderWerf, E. A. Prevalence of avian pox virus and effect on the fledging success of Laysan Albatross. J. Field Ornithol. 79, 93–98. https://doi.org/10.1111/j.1557-9263.2008.00149.x (2008).
van Riper, C., van Riper, S. G. & Hansen, W. R. Epizootiology and effect of avian pox on hawaiian forest birds. Auk 119, 929–942. https://doi.org/10.2307/4090224 (2002).
Walker, P. J. et al. Changes to virus taxonomy and the Statutes ratified by the international committee on taxonomy of viruses (2020). Arch Virol 165, 2737–2748. https://doi.org/10.1007/s00705-020-04752-x (2020).
Tulman, E. R. et al. The genome of Canarypox Virus. J Virol 78, 353–366. https://doi.org/10.1128/jvi.78.1.353-366.2004 (2004).
Offerman, K., Carulei, O., van der Walt, A. P., Douglass, N. & Williamson, A.-L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genom. 15, 1–17. https://doi.org/10.1186/1471-2164-15-463 (2014).
Banyai, K. et al. Unique genomic organization of a novel Avipoxvirus detected in turkey (Meleagris gallopavo). Infect. Genet. Evol. 35, 221–229. https://doi.org/10.1016/j.meegid.2015.08.001 (2015).
Afonso, C. L. et al. The genome of fowlpox virus. J. Virol. 74, 3815–3831 (2000).
Laidlaw, S. M. & Skinner, M. A. Comparison of the genome sequence of FP9, an attenuated, tissue culture-adapted European strain of Fowlpox virus, with those of virulent American and European viruses. J. Gen. Virol. 85, 305–322. https://doi.org/10.1099/vir.0.19568-0 (2004).
Joshi, L. R. et al. Detection of Fowlpox virus carrying distinct genome segments of Reticuloendotheliosis virus. Virus Res. 260, 53–59. https://doi.org/10.1016/j.virusres.2018.10.017 (2019).
Croville, G. et al. Rapid whole-genome based ty** and surveillance of avipoxviruses using nanopore sequencing. J. Virol. Methods 261, 34–39. https://doi.org/10.1016/j.jviromet.2018.08.003 (2018).
Sarker, S., Athukorala, A., Bowden, T. R. & Boyle, D. B. Characterisation of an Australian fowlpox virus carrying a near-full-length provirus of reticuloendotheliosis virus. Arch. Virol. 166, 1485–1488. https://doi.org/10.1007/s00705-021-05009-x (2021).
Sarker, S. et al. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.). BMC Genom. 18, 298. https://doi.org/10.1186/s12864-017-3680-z (2017).
Sarker, S. et al. Molecular characterisation of a novel pathogenic avipoxvirus from the Australian magpie (Gymnorhina tibicen). Virology 540, 1–16. https://doi.org/10.1016/j.virol.2019.11.005 (2020).
Sarker, S., Bowden, T. R. & Boyle, D. B. Genomic characterisation of a novel avipoxvirus, magpiepox virus 2, from an Australian magpie (Gymnorhina tibicen terraereginae). Virology 562, 121–127. https://doi.org/10.1016/j.virol.2021.07.010 (2021).
Sarker, S., Athukorala, A. & Raidal, S. R. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian passerine bird, mudlark (Grallina cyanoleuca). Virology 554, 66–74. https://doi.org/10.1016/j.virol.2020.12.011 (2021).
Sarker, S., Athukorala, A., Bowden, T. R. & Boyle, D. B. Genomic characterisation of a novel avipoxvirus isolated from an endangered yellow-eyed penguin (Megadyptes antipodes). Viruses 13, 194. https://doi.org/10.3390/v13020194 (2021).
Sarker, S., Bowden, T. R. & Boyle, D. B. Evidence of a possible viral host switch event in an avipoxvirus isolated from an endangered northern royal albatross (Diomedea sanfordi). Viruses 14, 302 (2022).
Sarker, S., Athukorala, A., Nyandowe, T., Bowden, T. R. & Boyle, D. B. Genomic characterisation of a novel avipoxvirus isolated from an endangered northern royal albatross (Diomedea sanfordi). Pathogens https://doi.org/10.3390/pathogens10050575 (2021).
McGraw, K., Penha, V. A. S., Drake, D., Kraberger, S. & Varsani, A. Poxvirus infection in house finches (Haemorhous mexicanus): Genome sequence analysis and patterns of infection in wild birds. Transbound Emerg. Dis. https://doi.org/10.1111/tbed.14575 (2022).
Grove, D. M., Zajac, A. M., Spahr, J., Duncan, R. B. Jr. & Sleeman, J. M. Combined infection by avian poxvirus and Collyriclum faba in an American crow (Corvus brachyrhynchos). J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 36, 111–114. https://doi.org/10.1638/03-100 (2005).
Gyuranecz, M. et al. Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 87, 4938–4951. https://doi.org/10.1128/JVI.03183-12 (2013).
Sarker, S. et al. Molecular and microscopic characterization of a novel Eastern grey kangaroopox virus genome directly from a clinical sample. Sci. Rep. 7, 16472. https://doi.org/10.1038/s41598-017-16775-7 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2 (1990).
Upton, C., Slack, S., Hunter, A. L., Ehlers, A. & Roper, R. L. Poxvirus orthologous clusters: Toward defining the minimum essential poxvirus genome. J. Virol. 77, 7590–7600. https://doi.org/10.1128/JVI.77.13.7590-7600.2003 (2003).
Lefkowitz, E. J., Wang, C. & Upton, C. Poxviruses: Past, present and future. Virus Res. 117, 105–118. https://doi.org/10.1016/j.virusres.2006.01.016 (2006).
Sarker, S., Isberg, S. R., Milic, N. L., Lock, P. & Helbig, K. J. Molecular characterization of the first saltwater crocodilepox virus genome sequences from the world’s largest living member of the Crocodylia. Sci. Rep. 8, 5623. https://doi.org/10.1038/s41598-018-23955-6 (2018).
Chung, Y. S. & Spradbrow, P. B. Studies on poxvirus isolated from a magpie in Queensland. Aust. Vet. J. 53, 334–336. https://doi.org/10.1111/j.1751-0813.1977.tb00243.x (1977).
Harrigan, K. E., Barker, I. K. & Studdert, M. J. Poxvirus infection in the white-backed magpie (Gymnorhina hypoleuca) and pox-like conditions in other birds in Australia. J. Wildl. Dis. 11, 343–347 (1975).
Mete, A., Borst, G. H. & Dorrestein, G. M. Atypical poxvirus lesions in two Galapagos doves (Nesopelia g. galapagoensis). Avian Pathol. 30, 159–162. https://doi.org/10.1080/03079450120044560 (2001).
Karstad, L. Pox. -. In Infectious and Parasitic Diseases of Wild Birds (eds Davis, J. W. et al.) 34–41 (Iowa State University Press, Iowa, 1971).
Forrester, D. J. The ecology and epizootiology of avian pox and malaria in wild turkeys. Bull. Soc. Vector Ecol. 16, 127–148 (1991).
Tripathy, D. N. et al. Characterization of poxviruses from forest birds in Hawaii. J. Wildl. Dis. 36, 225–230. https://doi.org/10.7589/0090-3558-36.2.225 (2000).
Lachish, S., Bonsall, M. B., Lawson, B., Cunningham, A. A. & Sheldon, B. C. Individual and population-level impacts of an emerging poxvirus disease in a wild population of great tits. PLoS ONE 7, e48545. https://doi.org/10.1371/journal.pone.0048545 (2012).
Sarker, S. et al. Crocodilepox virus evolutionary genomics supports observed poxvirus infection dynamics on saltwater crocodile (Crocodylus porosus). Viruses https://doi.org/10.3390/v11121116 (2019).
Sarker, S., Das, S., Helbig, K., Peters, A. & Raidal, S. R. Genome sequence of an Australian strain of canid alphaherpesvirus 1. Aust. Vet. J. 96, 24–27. https://doi.org/10.1111/avj.12659 (2018).
Sarker, S. Metagenomic detection and characterisation of multiple viruses in apparently healthy Australian Neophema birds. Sci. Rep. 11, 20915. https://doi.org/10.1038/s41598-021-00440-1 (2021).
Sarker, S. et al. Characterization of a complete genome sequence of molluscum contagiosum virus from an adult woman in Australia. Microbiol. Resour. Announc. https://doi.org/10.1128/mra.00939-20 (2021).
Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. https://doi.org/10.1006/jmbi.2000.4315 (2001).
Hofmann, K. & Stoffel, W. Tmbase-A database of membrane spanning protein segments. (1993).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a New HHpred server at its core. J. Mol. Biol. 430, 2237–2243. https://doi.org/10.1016/j.jmb.2017.12.007 (2018).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845. https://doi.org/10.1038/nprot.2015.053 (2015).
Waterhouse, A. et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296-w303. https://doi.org/10.1093/nar/gky427 (2018).
Maizel, J. V. Jr. & Lenk, R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl. Acad. Sci U.S.A. 78, 7665–7669 (1981).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. https://doi.org/10.1093/molbev/mst010 (2013).
Le Net, R. et al. Whole genome sequencing of an avipoxvirus associated with infections in a group of aviary-housed snow buntings (Plectrophenax nivalis). J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 50, 803–812. https://doi.org/10.1638/2018-0102 (2020).
Sarker, S., Hannon, C., Athukorala, A. & Bielefeldt-Ohmann, H. Emergence of a novel pathogenic poxvirus infection in the endangered green sea turtle (Chelonia mydas) highlights a key threatening process. Viruses 13, 219. https://doi.org/10.3390/v13020219 (2021).
Asif, K. et al. Whole-genome based strain identification of fowlpox virus directly from cutaneous tissue and propagated virus. PLoS ONE 16, e0261122. https://doi.org/10.1371/journal.pone.0261122 (2021).
Seitz, K. et al. Discovery of a phylogenetically distinct poxvirus in diseased Crocodilurus amazonicus (family Teiidae). Arch. Virol. 166, 1183–1191. https://doi.org/10.1007/s00705-021-04975-6 (2021).
Acknowledgements
Subir Sarker is the recipient of an Australian Research Council Discovery Early Career Researcher Award (Grant Number DE200100367) funded by the Australian Government. The Australian Government had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.S. and M.S.; Formal analysis, S.S.; Funding acquisition, S.S.; Investigation, S.S. and M.S.; Methodology, S.S. and M.S.; Writing—original draft, S.S.; Writing—review and editing, S.S. and M.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarker, S., Sutherland, M. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian little crow (Corvus bennetti) directly from the clinical sample. Sci Rep 12, 15053 (2022). https://doi.org/10.1038/s41598-022-19480-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19480-2
- Springer Nature Limited