Abstract
COVID-19 is a deadly outbreak that has been declared a public health emergency of international concern. The massive damage of the disease to public health, social life, and the global economy increases the importance of alternative rapid diagnosis and follow-up methods. RT-PCR assay, which is considered the gold standard in diagnosing the disease, is complicated, expensive, time-consuming, prone to contamination, and may give false-negative results. These drawbacks reinforce the trend toward medical imaging techniques such as computed tomography (CT). Typical visual signs such as ground-glass opacity (GGO) and consolidation of CT images allow for quantitative assessment of the disease. In this context, it is aimed at the segmentation of the infected lung CT images with the residual network-based DeepLabV3+, which is a redesigned convolutional neural network (CNN) model. In order to evaluate the robustness of the proposed model, three different segmentation tasks as Task-1, Task-2, and Task-3 were applied. Task-1 represents binary segmentation as lung (infected and non-infected tissues) and background. Task-2 represents multi-class segmentation as lung (non-infected tissue), COVID (GGO, consolidation, and pleural effusion irregularities are gathered under a single roof), and background. Finally, the segmentation in which each lesion type is considered as a separate class is defined as Task-3. COVID-19 imaging data for each segmentation task consists of 100 CT single-slice scans from over 40 diagnosed patients. The performance of the model was evaluated using Dice similarity coefficient (DSC), intersection over union (IoU), sensitivity, specificity, and accuracy by performing five-fold cross-validation. The average DSC performance for three different segmentation tasks was obtained as 0.98, 0.858, and 0.616, respectively. The experimental results demonstrate that the proposed method has robust performance and great potential in evaluating COVID-19 infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The novel coronavirus disease 2019 (COVID-19) was first reported in December 2019 in Wuhan City, Hubei province of China [1, 2]. Compared to the SARS-CoV epidemic in 2003, the contagiousness of COVID-19 was found to be more severe in a short period [2, 3]. The ability of COVID-19 to be transmitted from person to person, particularly through respiratory droplets and close contact, has led to an exponential increase in the number of positive cases [4]. COVID-19 was considered a public health emergency by the international community and declared a pandemic by the World Health Organization (WHO) on March 11, 2020. WHO confirms that there were approximately 236.5 million positive cases worldwide as of August 22, 2021. Unfortunately, 4,831,486 of the positive cases resulted in death [5]. The massive damage that the disease causes to public health, social life, and the global economy increase the importance of alternative rapid diagnostic and follow-up procedures.
The main procedure still used in the diagnosis of COVID-19 is based on laboratory tests. The reverse “ polymerase chain reaction (RT -PCR) assay is considered the gold standard for people with suspected COVID-19 who present to clinical centers with a variety of symptoms [6]. Although it is the main diagnostic kit, the method is time-consuming and leads to a considerable proportion of false-negative results [7, 8]. As an alternative to laboratory tests, chest X-ray and computed tomography (CT) scan have also proved helpful in diagnosing the disease and minimize the disadvantages of RT-PCR [3]. The fact that CT scans can be easily performed in most medical centers, have a high spatial resolution, and are more sensitive in detecting the irregularities in the lungs caused by COVID-19 encourages physicians to use this method frequently. However, the extraordinarily high number of positive cases poses a challenge to current screening methods. Considering the continuous analysis of radiological records routinely performed to monitor the course of the disease, this means a severe overload on the limited number of physicians [3, 9]. It is therefore believed that the systems to be developed to automatically detect and quantify lung infections caused by COVID-19 will help overburdened health centers manage the chaos well.
Visual symptoms of infections, such as ground-glass opacity (GGO) and consolidation, on CT images, have been confirmed to be major abnormalities in the diagnosis and evaluation of COVID-19. The incidences of GGO and consolidation were reported as 86% and 29%, respectively [10]. Changes in abnormalities over time contain crucial information about the disease. However, given the current caseload, manual review of changes in each patient may result in excessive time-consuming and human errors. Artificial intelligence (AI)-based investigation of CT scans can overcome the existing drawbacks. The AI approach can analyze disease-related lesions and various opacities faster and more quantitatively than the manual approach. The superior performance of deep learning algorithms in biomedical image processing in the last decade is the main motivation for this study.
The deep learning approach has achieved outstanding success, especially in recent years, in automatically extracting meaningful and unique features from existing medical datasets and classifying the extracted features [11, 12]. In these studies, the features extraction from the regions of interest and the diagnosis of the disease depending on the irregularities are used as a general procedure. However, deep learning-based semantic segmentation will provide more powerful solutions for tracking serious complications that may occur in parallel with the disease in the lungs. In the literature, semantic segmentation attempts are limited compared to the studies conducted for the detection of COVID-19 [13]. Semantic segmentation can be used to detect, spatially localize, and quantitatively evaluate disease-related lesions. This study aims to perform semantic segmentation of lung CT images in the fight against COVID-19 based on their tremendous potential.
Semantic segmentation is a method of interpreting an image at the pixel level. In other words, it is the process of assigning each pixel in the medical image to a predetermined class [14]. More specific information about diseases can be obtained by pixel-level analysis. With semantic segmentation, medical images can be understood not only for specific patterns but also by autonomous systems as a whole. In this way, a multidimensional analysis of lesions caused by COVID-19 in the lungs can be made possible. In this study, DeepLabV3+ architecture proposed in ref. [15] was applied for semantic segmentation. In the DeepLabV3+ architecture is a redesigned convolutional neural network (CNN) model for semantic segmentation. In the encoder structure of the segmentation model, a pre-trained ResNet-18 architecture is used as the backbone. ResNet-18 is an efficient network for applications with limited processing resources. Some of the significant and remarkable contributions of this study are listed below.
-
1.
The efficacy of residual network-based DeepLabv3+ has been demonstrated in the semantic segmentation of COVID-19 irregularities in CT images.
-
2.
Different segmentation tasks of chest CT images were examined and the factors affecting the segmentation performance of the regions of interest were investigated.
-
3.
The performances obtained in studies using common architectures in semantic segmentation of CT images were compared with the performance of the model proposed in this study.
-
4.
CT irregularities associated with COVID-19 infections were evaluated as separate classes, allowing the assessment of disease progression with more specific data.
Related work
Many successful models based on deep learning have been proposed in the literature for automatic detection and segmentation of infection irregularities. In studies of COVID-19 detection that propose popular CNN architectures by evaluating CT images as a whole, the problem of interest is generally considered binary (COVID-negative, COVID-positive) [16,17,18] or multi-class (normal, pneumonia, COVID) [8, 19]. Effective detection systems have also been developed using hybrid models where feature extraction is based on a deep learning approach and classification is based on conventional methods [20, 21]. In addition, state-of-the-art models based on transfer learning have been proposed due to the limited number of images related to the disease and the desire for higher classification accuracy [22,23,24].
The models proposed for detecting COVID-19 from radiographic scans evaluate the images as a whole and assign them to a specific class. These sophisticated systems work like RT -PCR diagnostic kits and reduce the burden on busy hospital clinics. However, a more in-depth analysis of CT symptoms associated with infections is needed to fight against COVID-19. It is well known that the nature and intensity of COVID-19 CT irregularities vary according to the course of the disease. Semantic segmentation can provide a comprehensive analysis of disease-related lung lesions. For this purpose, effective models for deep learning-based segmentation of COVID-19 lung lesions have been developed in the literature. The proposed models are mainly U-Net [25], SegNet [26], fully convolutional neural networks (FCN) [27] and optimized architectures based on these models. In this context, Amyar et al. [11] proposed a unique multi-task deep learning framework for the segmentation of COVID-19 lung lesions. To ensure the continuity of spatial information in CT images, they proposed a U-Net architecture in which they change the stride rate during the convolutional process instead of pooling in the encoder part. Zheng et al. [28] proposed a 3D CU -Net model based on 3D U-Net architecture for COVID -19 lesion segmentation. They propose an attention mechanism to achieve local cross-channel information interaction in an encoder to improve different levels of feature representation. Yan et al. [29] proposed a feature variation block that adaptively adjusts the global properties of features for the segmentation of COVID-19 infections. They combined features at different scales by proposing a progressive ASPP approach to supervise sophisticated infection areas with diverse appearances and shapes. Voulodimos et al. [30] analyzed the effectiveness of U-Net and FCN architectures in semantic segmentation of COVID -19 lesions. They proved that segmentation can be successfully performed despite the class imbalance in the dataset and human-made errors in labeling the boundaries of the symptom domains. Khalifa et al. [31] achieved an average IoU performance of 79.9% in semantic segmentation for a limited number of CT images by proposing a deep architecture consisting of an encoder and decoder structure. Saood et al. [13] performed semantic segmentation of COVID -19 lesions as binary and multi-class using SegNet and U-Net architectures and compared the performances of both models. Müller et al. [32] applied the standard 3D U-Net architecture to reduce the risk of overfitting in the segmentation problem where a limited number of images are used.
Materials and methods
In this study, an effective deep learning-based semantic segmentation model is proposed for the detection, localization, and quantitative evaluation of COVID-19 lung irregularities. The methods and dataset used following the defined objectives are introduced in detail.
Axial CT image database
The images in the dataset used in this study are a collection of the Italian Society of Medical and Interventional Radiology (SIRM) [33]. This dataset contains axial CT images from 68 patients with COVID-19. Most cases include multiple slices. Ultimately, 100 single-slice scans were presented from more than 40 patients to the researchers as 512 × 512 pixel grayscale images. Ground-truth maps of COVID-19 irregularities were prepared by an experienced radiologist. The radiologist labeled the infections at three levels: GGO (mask value=1), consolidation (=2), and pleural effusion (=3). In addition, lung masks are also available on the dataset resource site to contribute to computer-based automatic detection and segmentation models. Figure 1 shows an example of the original CT scan, lung mask, and ground-truth map of COVID-19 lesions.
Preprocessing for ground-truth maps
The segmentation process was applied for three different tasks to better analyze the effectiveness of the deep learning-based semantic segmentation architecture. Task-1 represents binary segmentation as lung (infected and non-infected tissue) and background. Task-2 represents multi-class segmentation as lung (non-infected tissue), COVID (GGO, consolidation, and pleural effusion irregularities are gathered under a single roof), and background. Finally, the segmentation in which each lesion type is considered as a separate class is defined as Task-3. In this context, the segmentation procedure was applied to the background, lung (non-infected tissue), GGO, consolidation, and pleural effusion classes. The ground-truth maps were rearranged without affecting the mask boundaries provided by the dataset source to implement various segmentation tasks. Figure 2 shows ground-truth maps for each segmentation task.
Ground-truth maps for different segmentation tasks. (a): Original axial CT scan, (b): Ground-truth map for Task-1 (black color is background and white is lung (infected and non-infected tissues) (c): Task-2 (black color is background, red is COVID-19 pneumonia, and white is lung (non-infected tissue), (d): Task-3 (black color is background, red is GGO, blue is consolidation, green is pleural effusion, and white is lung (non-infected tissue))
The details for deep learning-based semantic segmentation
In this section, the encoder and decoder structure commonly used in most state-of-the-art models for semantic segmentation is briefly introduced. The main components of the proposed DeepLabV3+ model, the dilated residual network that is the backbone structure, the atrous spatial pyramid pooling (ASPP), and finally the decoder module is explained. Figure 3 shows the dilated residual network-based DeepLabV3+ segmentation architecture used in the study. The proposed segmentation architecture was run separately on each segmentation task.
Encoder and decoder architecture
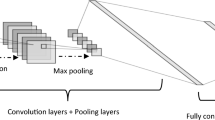
Most of the proposed methods for semantic segmentation consist of encoder and decoder blocks [13]. The image presented to the input in the encoder section is progressively downsampled. The resolution of the feature maps is gradually decreased to capture the high-level information. The generation of the segmentation map with the same size as the original image from the small size feature maps supplied by the encoder is achieved with an efficient decoder module. With the decoder, in which spatial information is captured, a faster and stronger overall architecture is created [14, 15]. A general encoder and decoder structure based on deep learning applied in semantic segmentation is shown in Fig. 4. In this study, the dilated residual network (ResNet) was used as the backbone in the encoder block.
Dilated residual network for encoder
Deep learning outperforms conventional methods, especially in classifying large-scale images [34, 35]. Recently, many popular CNN architectures have been proposed that include various processing units and topological differences. It was observed that although the training process becomes increasingly difficult, the performance does not increase proportionally with depth [36, 37]. This is mainly because of the vanishing and exploding gradient problem [35]. To facilitate the training of networks and pave the way for deeper architectures, residual blocks were introduced in ref. [36]. Thus, it has been shown that effective training of deeper networks can increase accuracy with increasing depth. ResNet architectures are a popular feature extractor in semantic segmentation tasks, which has a lower computational cost despite its deeper architecture. In this study, the dilated ResNet architecture is used as the backbone structure for the semantic segmentation model to achieve high segmentation accuracy. Although the number of layers is various such as ResNet-18, ResNet-34, and ResNet-50, the networks have a basic form. ResNet uses five convolution blocks with output spatial resolution. Therefore, it has been found sufficient to use the simplest of these architectures.
In the ResNet-18 structure, where the residual block is the base element of each convolutional block, the small-scale feature maps generated after the encoder have a negative impact on the segmentation performance. The stride rate is set to 1 instead of 2 in the convolutional layer in the last two blocks of the architecture to obtain feature maps that reflect high spatial information. However, with the reduction of the stride factor, the loss of receptive field in the convolution process was tried to be compensated by the appropriate dilation factor. Dilation rate (d=1) represents standard convolution operation, d>1 represents upsampled convolution filters. Figure 5 shows dilated convolution kernels at different d rates.
Depending on the determined d rates, a balance between the feature map and the receptive field is achieved with dilation convolutions, where zeros are placed between the filter weights. The size of the 16 × 16 final feature maps obtained for a 512 × 512 pixels image with the original ResNet architecture can be increased by dilated convolution. The typical downsampling factor (also called Output Stride (OS)) in CNN is 32. OS represents the ratio of spatial resolution of the input image and output image [15]. With the dilation approach, the OS ratio was set to 16 in this study. Attempts were made to obtain stronger segmentation maps by achieving more intensive feature extraction at a lower OS rate.
Atrous spatial pyramid pooling (ASPP)
The ASPP technique was developed with the motivation of the success of atrous convolutional operations and spatial pyramid pooling (SPP). The SPP was developed to overcome the requirement of a fixed size for the input images in deep learning [38]. ASPP technique resamples feature maps generated from the encoder at different atrous rates. Then, the outputs obtained by applying a parallel convolution filter to the feature maps at different atrous rates are concatenated as shown in Fig. 3. Although extensive semantic information is encoded in the final feature map, boundary information may be lost for the region of interest [15]. Multi-scale information can be captured accurately and effectively with the ASPP method [14]. In this study, the ASPP module that consists of 1 × 1 convolution, followed by 3 × 3 convolutions with dilation rate d= 6, 12, and 18 and max-pooling layer in parallel was used. With depth-wise convolution applied instead of standard convolution, COVID-19 irregularities of various densities and sizes have been trying to be segmented with high sensitivity.
Decoder module
The decoder module generates segmentation maps in original image size from low-scale feature maps of the encoder. The encoder feature map is downsampled by OS=16 than the original image. As seen in Fig. 3, the encoder feature maps are first upsampled with factor 4 to produce the semantic segmentation map. These feature maps are concatenated with intermediate low-level features from the encoder backbone with the same spatial dimension. Segmentation performance is improved by concatenating low-level features rich in spatial information from the backbone network and high-level features from ASPP [15, 33]. Then, 3 × 3 convolution is performed before this feature map is again upsampled by 4 to obtain the final segmentation output.
Training procedure and hyperparameters tuning
The preprocessing steps and segmentation experiments were performed in a Matlab programming environment (The MathWorks, Natick, MA, USA) installed on a 2.7 GHz dual Intel i7 processor 16 GB RAM, NVIDIA GeForce ROG-STRIX 256 bit, and 8GB GPU hardware.
The one-hundred images dataset is divided into three sets for training, validation, and testing, with proportions of 60%, 20%, and 20% respectively. In order to reliably evaluate the segmentation performance, five-fold cross-validation was performed for all segmentation tasks. In this way, the ability of the residual network-based DeepLabV3+ to achieve semantic segmentation of COVID-19 lesions was evaluated. Training of network is performed with the ADAM stochastic optimizer because it has a fast convergence rate compared to other optimizers [39]. Rectified Linear Unit (ReLU) was used after each depth separable convolution. ReLU is an activation function, which has robust biological and mathematical underpinning. To improve the performance of the segmentation model, various hyperparameters such as initial learning rate, mini-batch size, and maximum epoch were tuned to an optimal level through trial and error. In the training of the network, the appropriate initial learning rate was determined as 0.0001 to avoid a long period in which the network can get stuck during learning or rapid learning without optimal weight values. For the proposed model to perform successful learning, mini-batch size and maximum epoch number were set to 4 and 10, respectively.
Evaluation metrics
The performance of any semantic segmentation model can only be reliably evaluated if appropriate metrics are considered. Segmentation of medical images is generally between unbalanced classes in terms of pixel distribution. In medical image segmentation, such as COVID-19 infections on CT, the region of interest is too small compared to the entire image and is therefore represented by fewer true positives (TP). True negatives representing background or lung tissue (non-infected) may result in misleading performance ratings for accuracy due to class imbalances. Therefore, it is necessary to focus on the Dice similarity coefficient (DSC) and intersection over union (IoU) (also known as Jaccard index) metrics based on F-score that robustly and reliably reflect model performance [32]. In this study, the performance of the proposed DeepLabV3+ based CT segmentation model was evaluated in terms of these metrics. In addition, accuracy, sensitivity and specificity metrics that are commonly used in the literature are also given.
Model metrics are based on the confusion matrix for binary classification, where TP, FP, TN and FN represent the true positive, false positive, true negative and false negative rate, respectively. The statistical measures used to evaluate the performance were:
TP: the number of pixels correctly associated with the region of interest.
TN: the number of pixels correctly associated with the class outside the region of interest.
FP: the pixels associated with the region of interest by the proposed model but not by the radiologist.
FN: the pixels associated with the region of interest by the radiologist but missed by the proposed model.
Results
Binary segmentation (Task-1)
The binary segmentation experiment of lung CT images as background and lung (infected and non-infected tissue) was first performed to evaluate the effectiveness of the proposed model. The effect of distribution between classes on semantic segmentation performance has been tried to be analyzed. Since infected and non-infected tissues were labeled as lung, thus achieving a more balanced distribution between classes according to other segmentation tasks. Table 1 shows the pixel count (total number of pixels in a class) and image pixel count (total number of pixels in images that had an instance of the class) information regarding lung tissue and background classes.
Following training, inference revealed strong segmentation performance for lungs and background region. As a result of the five-fold cross-validation, the proposed model achieved average segmentation performance with IoU of 0.942 and 0.979 for lung and background classes, respectively. In addition, the DSC performance for both classes was 0.971 and 0.989, respectively. The results show that the segmentation model can discriminate the lung tissue on axial CT images. Overall, the proposed model achieved a DSC of 0.98 for all classes. Furthermore, the models obtained a sensitivity and specificity of 0.977 and 0.987 on the testing set for lung tissue, respectively. Table 2 shows the details regarding the performance exhibited by the proposed model for binary segmentation.
Multi-class segmentation (Task-2)
GGO, consolidation, and pleural effusion irregularities were gathered under one roof in the segmentation process and accepted as a collective pattern in the COVID-19 detection. The proposed model has attempted to semantically segment irregularities labeled as COVID. As a result, the segmentation process was applied for the classes related to background, lung (non-infected tissue), and COVID. Table 3 shows the pixel count and image pixel count data for COVID, lung (non-infected tissue), and background classes.
As can be seen in Table 3, although three different COVID-19 irregularities were considered as one pattern, the distributional imbalance between classes increased according to Task-1 segmentation. The segmentation model obtained an average DSC and IoU of 0.858 and 0.772, respectively. Overall, the 5 fold cross-validation process resulted by DSC and IoU of 0.882 and 0.789 for the lungs (non-infected tissue), as well as 0.708 and 0.548 for COVID-19 infection. For background class segmentation, DSC and IoU metrics were obtained as 0.985 and 0.972, respectively. Overall, the proposed model achieved a DSC of 0.858 for all classes. The proposed model was able to segment infected and non-infected lung tissues successfully. Sensitivity performance was 0.792, 0.866, and 0.979 for COVID-19 lesions, lung (non-infected tissues), and background classes, respectively. The sensitivity and specificity results show the effectiveness of false negative decisions on model performance due to unbalanced distribution between classes (Table 4).
Multi-class segmentation (Task-3)
A detailed segmentation process based on COVID-19 irregularities types was performed. For this purpose, GGO, consolidation, and pleural effusion irregularities were considered separate classes. Considering the COVID-19 irregularities separately causes the focused regions to be smaller and thus the imbalance in the pixel distribution between classes increases in parallel. Table 5 shows the pixel count and image pixel count data of the classes considered in the segmentation.
It is known that CT symptoms progress in different intensities in parallel with the lung destruction of COVID-19. Table 5 confirms from the pixel count data that COVID-19 irregularities are frequently seen as GGO and consolidation in the dataset. It can be seen that pleural effusion is observed much less frequently compared to other findings and is not included in every image. So it is clear that false negative decisions for pleural effusion are much more effective than other classes. The unbalanced segmentation problem also leads to a high level of variance in the performance obtained for each fold during the training phase. Table 6 shows the performance metrics of the proposed model for Task-3 segmentation in detail.
The proposed model achieved DSC performances of 0.597 and 0.511 for GGO and consolidation CT irregularities, respectively. For lung class segmentation, average DSC were obtained as 0.881. It can be said that the performance is low due to the very few pleural effusion patterns and false negatives sabotage the performance. However, it has been observed that the overall performance is quite reasonable for the multi-class segmentation. Overall, the proposed model achieved a DSC of 0.616 for all classes. In general, it is clear from the standard deviation values that the proposed model shows stable performance for different folds in the training phase on various segmentation tasks. It can be seen that the presence of different CT samples in each training and testing phase does not introduce risky variations in model performance. Figure 6 shows the training accuracy and loss functions for each fold related to different segmentation tasks.
The results proved that the fluctuations in training accuracy and loss functions increase in direct proportion to the number of classes in the segmentation tasks. In particular, it was observed that the training process was stopped early for some folds in order not to be overfitting. Figure 7 shows the visual results obtained for some CT image examples in different segmentation tasks.
Discussion
The CT method is one of the most powerful tools in the diagnosis and evaluation of COVID-19. Compared to the classification-based models proposed for COVID-19 detection, the semantic segmentation approach provides detection, spatial localization, and quantitative evaluation of infections. In this regard, it is clear that the semantic segmentation method provides rich and robust information about the disease. The small size of the data set used may negatively affect the training from scratch of the backbone architecture used in the encoder section. Therefore, the pre-trained architecture has been successful in producing rich feature maps in the Large-Scale Visual Recognition Challenge (ILSVRC). The proposed model presented optimal segmentation maps by concatenating low-level features and high-level features. The main problem in segmentation performance is the dominance of false negatives since the regions of interest are quite small compared to the whole image. The very few false positive and false negative results may cause a tremendous impact on the metric values. The decrease in performance from larger to smaller regions of interest is evidenced by achieving an average DSC of 0.98, 0.858, and 0.616 for Task-1, Task-2, and Task-3 segmentation operations, respectively. Moreover, the lower level of performance for COVID-19 irregularities compared to other segmentation tasks is also due to the morphological diversity of lesions.
From the literature reviewed, it can be seen that several successful models have been proposed for segmenting the characteristic irregularities caused by COVID-19 infections in lung CT images. The studies are mostly based on U-Net, SegNet, and FCN architectures and various modifications made to these architectures. Table 7 compares the performances of the proposed state-of-the-art models for semantic segmentation of COVID-19 infections and the DeepLabV3+ model used in this study. The performance of the segmentation model was evaluated in terms of accuracy, sensitivity, specificity, DSC, and IoU metrics. The accuracy metric may result in misleading performance ratings due to class imbalances. Thus, the science community focused mainly on the other four widely popular metrics for medical image analysis.
Segmentation of COVID-19 irregularities from CT images is a crucial step for identifying and quantifying disease. Autonomous systems developed in this context allow effective monitoring of the course of the disease. Thus, many state-of-the-art models have been conducted to improve segmentation performance. Table 7 presents the segmentation performance of encoder decoder-based popular CNN architectures. All state-of-the-art models can achieve efficient performance for the segmentation of COVID-19 lesions. However, segmentation approaches have a high number of parameters, computing time, and network complexity. These drawbacks can make them infeasible for real-world applications. Various backbone options can make the DeepLabV3+ model smaller than other architectures.
The main challenges of COVID-19 irregularities segmentation are the small size of lesions, anatomical differences, and human-made errors in labeling the boundaries. In this study, although the convolution and pooling operations in the encoder part provide rich information, the boundary information about the regions of interest may be lost. To increase resolution, the downsampling or strided convolution can be removed from the network architecture. But it has the major drawback of reduction in the receptive field of kernels. It is main challenging to establish a balance between the resolution of the feature map and the receptive field. These challenges have been tried to be overcome with various improvements such as dilation convolution operation and different atrous rates.
DeeplabV3+was originally developed for the segmentation of road scenes, biomedical and real-world images. It generally gives better performance for various segmentation problems compared to FCN, SegNet, and U-Net architectures [14, 43, 44]. However, DeepLabV3+ is considered relatively unexplored in the analysis of CT images for COVID-19. The proposed method performed similar or significantly better segmentation performance according to related works without increasing the number of model parameters by using atrous convolution and SPP methods. Thus, it has been shown that the DeepLabV3+ architecture may be one of the main approaches used in the segmentation of COVID-19 lesions.
Conclusions
In this study, a multi-task semantic segmentation was performed for the detection of COVID-19 pulmonary infections. DeepLabV3+ architecture, a recently developed CNN model, was used instead of the segmentation models commonly used in the literature. A pre-trained ResNet-18 architecture was used as a backbone to improve the feature representation capability. The effectiveness of the proposed DeepLabV3+ was evaluated using the segmentation results for healthy lung tissue and different types of COVID-19 lesions. The experimental results demonstrate that the proposed method has robust performance and great potential in evaluating COVID-19 infection.
The segmentation performance can be improved via further research on various parameter modifications of the DeepLabV3+. In particular, different base networks can be employed for feature extractor backbone. In addition, the DeepLabV3+ framework may work more effectively by using different atrous rates in the encoder or ASPP module.
References
Toğaçar M, Ergen B, Cömert Z (2020) COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 121: https://doi.org/10.1016/j.compbiomed.2020.103805.
Pereira RM, Bertolini D, Teixeira LO et al (2020) COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed 194. https://doi.org/10.1016/j.cmpb.2020.105532
Rohila VS, Gupta N, Kaul A, Sharma DK (2021) Deep learning assisted COVID-19 detection using full CT-scans. Internet of Things (Netherlands) 14:100377. https://doi.org/10.1016/j.iot.2021.100377
Loey M, Manogaran G, Taha MHN, Khalifa NEM (2021) A hybrid deep transfer learning model with machine learning methods for face mask detection in the era of the COVID-19 pandemic. Meas J Int Meas Confed 167:108288. https://doi.org/10.1016/j.measurement.2020.108288
Coronavirus disease (COVID-19). https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 9 Oct 2021
Xu X, Jiang X, Ma C et al (2020) A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering 6:1122–1129. https://doi.org/10.1016/j.eng.2020.04.010
Oulefki A, Agaian S, Trongtirakul T, Kassah Laouar A (2021) Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit 114:107747. https://doi.org/10.1016/j.patcog.2020.10774
Mishra NK, Singh P, Joshi SD (2021) Automated detection of COVID-19 from CT scan using convolutional neural network. Biocybern Biomed Eng 41:572–588. https://doi.org/10.1016/j.bbe.2021.04.006
Desai SB, Pareek A, Lungren MP (2020) Deep learning and its role in COVID-19 medical imaging. Intell Med 3–4:100013. https://doi.org/10.1016/j.ibmed.2020.100013
Li M (2020) Chest CT features and their role in COVID-19. Radiol Infect Dis 7:51–54. https://doi.org/10.1016/j.jrid.2020.04.001
Amyar A, Modzelewski R, Li H, Ruan S (2020) Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Comput Biol Med 126:104037. https://doi.org/10.1016/j.compbiomed.2020.104037
Benameur N, Mahmoudi R, Zaid S et al (2021) SARS-CoV-2 diagnosis using medical imaging techniques and artificial intelligence: A review. Clin Imaging 76:6–14. https://doi.org/10.1016/j.clinimag.2021.01.019
Saood A, Hatem I (2021) COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med Imaging 21:1–11. https://doi.org/10.1186/s12880-020-00529-5
Baheti B, Innani S, Gajre S, Talbar S (2020) Semantic scene segmentation in unstructured environment with modified DeepLabV3+. Pattern Recognit Lett 138:223–229. https://doi.org/10.1016/j.patrec.2020.07.029
Chen LC, Zhu Y, Papandreou G et al (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 11211 LNCS:833–851. https://doi.org/10.1007/978-3-030-01234-2_49
Sethy PK, Behera SK, Ratha PK, Biswas P (2020) Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci 5:643–651. https://doi.org/10.33889/IJMEMS.2020.5.4.052
Hemdan EED, Shouman MA, Karar ME (2020) COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. ar**v preprint ar**v:2003.11055
Polat H, Özerdem MS, Ekici F, Akpolat V (2021) Automatic detection and localization of COVID-19 pneumonia using axial computed tomography images and deep convolutional neural networks. Int J Imaging Syst Technol 31:509–524. https://doi.org/10.1002/ima.22558
JavadiMoghaddam SM, Gholamalinejad H (2021) A novel deep learning based method for COVID-19 detection from CT image. Biomed Signal Process Control 70:102987. https://doi.org/10.1016/j.bspc.2021.102987
Kassania SH, Kassanib PH, Wesolowskic MJ et al (2021) Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning Based Approach. Biocybern Biomed Eng 41:867–879. https://doi.org/10.1016/j.bbe.2021.05.013
Ohata EF, Bezerra GM, Chagas JVS, Das et al (2021) Automatic detection of COVID-19 infection using chest X-ray images through transfer learning. IEEE/CAA J Autom Sin 8:239–248. https://doi.org/10.1109/JAS.2020.1003393
Minaee S, Kafieh R, Sonka M et al (2020) Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal 65. https://doi.org/10.1016/j.media.2020.101794
Taresh MM, Zhu N, Ali TAA et al (2021) Transfer Learning to Detect COVID-19 Automatically from X-Ray Images Using Convolutional Neural Networks. Int J Biomed Imaging 2021:. https://doi.org/10.1155/2021/8828404
Li C, Yang Y, Liang H, Wu B (2021) Transfer learning for establishment of recognition of COVID-19 on CT imaging using small-sized training datasets[Formula presented]. Knowledge-Based Syst 218:106849. https://doi.org/10.1016/j.knosys.2021.106849
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 9351:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39:2481–2495. https://arxiv.org/pdf/1511.00561.pdf
Shelhamer E, Long J, Darrell T (2017) Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell 39:640–651. https://doi.org/10.1109/TPAMI.2016.2572683
Zheng R, Zheng Y, Dong-Ye C (2021) Improved 3D U-Net for COVID-19 Chest CT Image Segmentation. https://doi.org/10.1155/2021/9999368. Sci Program 2021:
Yan Q, Wang B, Gong D et al (2020) COVID-19 Chest CT Image Segmentation – A Deep Convolutional Neural Network Solution. ar**v preprint ar**v:2004.10987
Voulodimos A, Protopapadakis E, Katsamenis I et al (2021) Deep learning models for COVID-19 infected area segmentation in CT images. ACM Int Conf Proceeding Ser 404–411. https://doi.org/10.1145/3453892.3461322
Khalifa NEM, Manogaran G, Taha MHN, Loey M (2021) A deep learning semantic segmentation architecture for COVID-19 lesions discovery in limited chest CT datasets. Expert Syst 1–11. https://doi.org/10.1111/exsy.12742
Müller D, Soto-Rey I, Kramer F (2021) Robust chest CT image segmentation of COVID-19 lung infection based on limited data. Inf Med Unlocked 25. https://doi.org/10.1016/j.imu.2021.100681
COVID-19 - Medical segmentation. http://medicalsegmentation.com/covid19/. Accessed 10 Oct 2021
Jia Deng W, Dong, Socher R et al (2009) ImageNet: A large-scale hierarchical image database. 248–255. https://doi.org/10.1109/cvprw.2009.5206848
Baheti B, Gajre S, Talbar S (2019) Semantic Scene Understanding in Unstructured Environment with Deep Convolutional Neural Network. IEEE Reg 10 Annu Int Conf Proceedings/TENCON 2019-Octob:790–795. https://doi.org/10.1109/TENCON.2019.8929376
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016-Decem 770–778. https://doi.org/10.1109/CVPR.2016.90
Sarwinda D, Paradisa RH, Bustamam A, Anggia P (2021) Deep Learning in Image Classification using Residual Network (ResNet) Variants for Detection of Colorectal Cancer. Procedia Comput Sci 179:423–431. https://doi.org/10.1016/j.procs.2021.01.025
He K, Zhang X, Ren S, Sun J (2014) Spatial pyramid pooling in deep convolutional networks for visual recognition. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 8691 LNCS:346–361. https://doi.org/10.1007/978-3-319-10578-9_23
Kingma DP, Ba JL (2015) Adam: A method for stochastic optimization. 3rd Int Conf Learn Represent ICLR 2015 -Conf Track Proc, 1–15
Jun M, Cheng G, Yixin W et al (2020) COVID-19 CT Lung and Infection Segmentation Dataset. https://doi.org/10.5281/ZENODO.3757476
Ma J, Wang Y, An X et al (2021) Toward data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation. Med Phys 48:1197–1210. https://doi.org/10.1002/mp.14676
Pei HY, Yang D, Liu GR, Lu T (2021) MPS-net: Multi-point supervised network for ct image segmentation of covid-19. IEEE Access 9:47144–47153. https://doi.org/10.1109/ACCESS.2021.3067047
Khan Z, Yahya N, Alsaih K et al (2020) Evaluation of deep neural networks for semantic segmentation of prostate in T2W MRI. Sens (Switzerland) 20:1–17. https://doi.org/10.3390/s20113183
Menteşe E, Hançer E (2020) Nucleus segmentation with deep learning approaches on histopathology images. Eur J Sci Technol 95–102. https://doi.org/10.31590/ejosat.819409
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Declarations
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Polat, H. Multi-task semantic segmentation of CT images for COVID-19 infections using DeepLabV3+ based on dilated residual network. Phys Eng Sci Med 45, 443–455 (2022). https://doi.org/10.1007/s13246-022-01110-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-022-01110-w