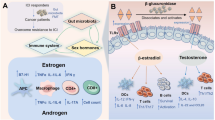
Abstract
The prevalence and progression of cancer differ in males and females, and thus, sexual dimorphism in tumor development directly impacts clinical research and medicine. Long non-coding RNAs (lncRNAs) are increasingly recognized as important players in gene expression and various cellular processes, including cancer development and progression. In recent years, lncRNAs have been implicated in the differences observed in cancer incidence, progression, and treatment responses between men and women. Here, we present a brief overview of the current knowledge regarding the role of lncRNAs in cancer sex dimorphism, focusing on how they affect epigenetic processes in male and female mammalian cells. We discuss the potential mechanisms by which lncRNAs may contribute to sex differences in cancer, including transcriptional control of sex chromosomes, hormonal signaling pathways, and immune responses. We also propose strategies for studying lncRNA functions in cancer sex dimorphism. Furthermore, we emphasize the importance of considering sex as a biological variable in cancer research and the need to investigate the role lncRNAs play in mediating these sex differences. In summary, we highlight the emerging link between lncRNAs and cancer sex dimorphism and their potential as therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The incidence of cancer, therapeutic response, and overall survival of cancer patients differ between men and women (Henley et al. 2020; Islami et al. 2021). Males are generally more likely to develop cancer. The National Cancer Institute reported through the Surveillance Epidemiology and End Results (SEER) database that for all cancers combined in the United States from 2016 to 2020, the age-adjusted incidence rate (and 95% confidence interval) per 100,000 was 477.8 ± 0.7 for males and 412.8 ± 0.6 for females. The same data analysis for the cancer mortality rate was 177.5 ± 0.3 for males and 128.7 ± 0.2 for females. The sex disparity in cancer is not restricted to any country or region. For the year 2020, the Global Cancer Observatory GLOBOCAN database provided statistics for 36 cancer types across 185 countries, with an age-adjusted incidence rate of 222.0 for males and 186.0 for females. In 2020, the world’s cancer mortality rate was 120.8 for males and 84.2 for females. We present in Table 1 the incidence rates for major cancers based on sex. Males have higher incidence rates for many types of cancer.
The numbers reported in Table 1 include sex-specific reproductive cancer types, such as ovarian and prostate cancers. The common non-reproductive cancers include the types that have high male-to-female ratios: colorectal cancers, lung and liver, and non-Hodgkin lymphoma. The analysis of SEER 2020 (USA) identified Kaposi sarcoma as having the highest male-to-female incidence rate ratio. In addition to breast cancer, which is rare in males, only a few cancers are more common in females, which is similar to what was previously noted (Dorak and Karpuzoglu 2012). In the case of gallbladder and thyroid cancer types, the male-to-female incidence rate ratio is less than 1.0. Although lifestyles are known to contribute to these differences, genetics also play an important role, and the molecular mechanisms involved are largely unknown. Despite traditional beliefs that sex hormones and hormonal regulation are the main explanatory factors for sexual dimorphism in cancer, accumulating evidence suggests that additional genetic and epigenetic mechanisms are at play. However, the specific molecular and cellular mechanisms responsible for sexual dimorphisms in cancer incidence and therapeutic responses are still in the early stages of discovery (Sandovici et al. 2022).
Non-coding RNAs (ncRNAs; RNAs that do not encode proteins) play critical roles in gene regulation (Mattick and Makunin 2006; Nair et al. 2020). Long noncoding RNAs (lncRNAs) are larger than 200 nucleotides and are important building blocks of gene regulatory networks in all eukaryotes (Kopp and Mendell 2018; Rinn and Chang 2012; Yao et al. 2019). LncRNAs share many similarities to messenger RNA (mRNA), including transcription by RNA Polymerase II, and undergoing co- and post-transcriptional processing events such as splicing, 5′ cap**, and 3′ polyadenylation (Cuykendall et al. 2017; Mattick et al. 2023; Quinn and Chang 2016). The human genome encodes thousands of lncRNAs, which represent potentially key sources of gene regulatory adaptation (Djebali et al. 2012). Analyzing lncRNAs is challenging as they are often expressed at low levels. Some investigators consider lncRNAs to be transcriptional “noise”. Therefore, the functions for most lncRNAs remain obscure and their biological importance is disputed. With advances in whole-genome technologies, increasing research on lncRNAs has highlighted their important role in a wide range of cellular processes. LncRNAs participate in chromatin remodeling and transcription, splicing, translation, and processing, localizing, and stabilizing other RNAs (Mattick et al. 2023).
An important role for lncRNAs in tumor initiation and progression has been established in recent years (Bhan et al. 2017; Jiang et al. 2019). Their tissue-specific expression makes them attractive for diagnostic and therapeutic purposes. For instance, the lncRNA PCA3 (Prostate Cancer Antigen 3) is used as a diagnostic marker for prostate cancer, and it can be easily found in urine samples (Taheri et al. 2022); the lncRNA HOTAIR (HOX antisense intergenic RNA) is involved in hormone therapies resistant in breast cancer (Xue et al. 2021; Syrett et al. 2019; Viggiano et al. 2016; Yu et al. 2021). Age-acquired skewed X-inactivation has recently been linked to cancer incidence (Roberts et al. 2022).
X-inactivation is an epigenetic hallmark of mammalian development (Fang et al. 2019; Payer et al. 2011). Two X-chromosomes in a female cell activate the expression of the X-Inactivation-Specific Transcript (XIST). In females, XIST is an lncRNA of 17-kb long that causes transcriptional inactivation of one of the two X-chromosomes, and this effect persists in all somatic cells throughout their life. LncRNA XIST and its flanking regulatory lncRNAs genes such as JPX (Just Proximal of ** (SA) can be induced, thereby disrupting the targeted lncRNA’s function (Liu et al. 2017). Large genomic screens can also be performed using Antisense Oligo Nucleotide (ASO) and RNA interference (RNAi) (Yip et al. 2022). To identify cancer sex-specific lncRNAs, a screen in female and male cancer cell lines must be performed. A variety of phenotypes can be asserted, such as survival, proliferation, invasion, and migration. A major advantage of the genetic screen approach is the ability to identify genes involved in tumor progression and compare their impact on female and male cells. Using cell lines has the disadvantage of missing all aspects of the tumor microenvironment (TME) that could impact sex-specific gene regulation. Therefore, clinical validation of the findings is essential.
Summary
Personalized therapies make it increasingly important to take biological sex into account. This review shows that lncRNAs play a role in cancer progression with different outcomes based on the sex. The molecular mechanisms behind this dimorphism need to be understood. LncRNAs are promising therapeutic targets that can also be used as diagnostic tools. There are several pathways involving lncRNAs, including chromosome X inactivation, hormonal signaling, and immunity. However, it is important to note that these are all interconnected pathways. Indeed, it is well known that the immune system is dependent on the signaling of sex hormones’ signaling, and that some immune genes are expressed from the X chromosome, including TLR8 (Toll-like receptor 8), TLR7 (Toll-like receptor 7), and IRAK1 (Interleukin-receptor-associated kinase 1). Moreover, it is difficult to determine how lifestyle influences cancer sex disparities and how environmental factors interact with sex-specific lncRNAs in affecting cancer. For the lncRNAs highlighted in this review, however, further investigation is required to establish a causal relationship with cancer sex dimorphism. The identification of the molecular mechanisms underlying cancer sex dimorphism may lead to the development of more targeted and effective cancer treatment options for males and females. In addition to its scientific impact, the characterization of sex dimorphism-related lncRNA could also have a positive impact on communities disproportionately at risk or afflicted by cancer. As a result of improving cancer treatment, we could potentially reduce cancer-related health disparities and improve health outcomes overall.
References
Abdel-Hafiz HA, Schafer JM, Chen X, **ao T, Gauntner TD, Li Z, Theodorescu D (2023) Y chromosome loss in cancer drives growth by evasion of adaptive immunity. Nature 619(7970):624–631. https://doi.org/10.1038/s41586-023-06234-x
Arun G, Diermeier S, Akerman M, Chang K-C, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL (2016) Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30(1):34–51. https://doi.org/10.1101/gad.270959.115
Bhan A, Hussain I, Ansari KI, Bobzean SAM, Perrotti LI, Mandal SS (2014) Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol 141:160–170. https://doi.org/10.1016/j.jsbmb.2014.02.002
Bhan A, Soleimani M, Mandal SS (2017) Long non-coding RNA (LncRNA) and cancer: a new paradigm. Cancer Res 77(15):3965–3981. https://doi.org/10.1158/0008-5472.CAN-16-2634
Brockdorff N, Bowness JS, Wei G (2020) Progress toward understanding chromosome silencing by **st RNA. Genes Dev 34(11–12):733–744. https://doi.org/10.1101/gad.337196.120
Castro A, Pyke RM, Zhang X, Thompson WK, Day C-P, Alexandrov LB, Zanetti M, Carter H (2020) Strength of immune selection in tumors varies with sex and age. Nat Commun 11(1):4128. https://doi.org/10.1038/s41467-020-17981-0
Chaligné R, Popova T, Mendoza-Parra M-A, Saleem M-AM, Gentien D, Ban K, Piolot T, Leroy O, Mariani O, Gronemeyer H, Vincent-Salomon A, Stern M-H, Heard E (2015) The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res 25(4):488–503. https://doi.org/10.1101/gr.185926.114
Chen Y, Sun Y, Rao Q, Hua X, Li L, Chang C (2015) Androgen receptor (AR) suppresses miRNA-145 to promote renal cell carcinoma (RCC) progression independent of VHL status. Oncotarget 6(31):31203–31215. https://doi.org/10.18632/oncotarget.4522
Chou K, Henderson J (2014) Endocrine system. In: Wexler P (ed) Encyclopedia of toxicology, 3rd edn. Academic Press, New York, pp 332–340. https://doi.org/10.1016/B978-0-12-386454-3.00377-8
Clusan L, Ferrière F, Flouriot G, Pakdel F (2023) A basic review on estrogen receptor signaling pathways in breast cancer. Int J Mol Sci 24(7):6834. https://doi.org/10.3390/ijms24076834
Conforti F, Pala L, Pagan E, Bagnardi V, De Pas T, Queirolo P, Pennacchioli E, Catania C, Cocorocchio E, Ferrucci PF, Saponara M, Orsolini G, Zagami P, Nicoló E, De Marinis F, Tortora G, Bria E, Minucci S, Joffe H et al (2021) Sex-based dimorphism of anticancer immune response and molecular mechanisms of immune evasion. Clin Cancer Res 27(15):4311–4324. https://doi.org/10.1158/1078-0432.CCR-21-0136
Cuykendall TN, Rubin MA, Khurana E (2017) Non-coding genetic variation in cancer. Curr Opin Syst Biol 1:9–15. https://doi.org/10.1016/j.coisb.2016.12.017
Ding J, Yeh C-R, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S (2018) Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 37(37):5037–5053. https://doi.org/10.1038/s41388-018-0175-6
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Gingeras TR (2012) Landscape of transcription in human cells. Nature 489:7414. https://doi.org/10.1038/nature11233
Dorak MT, Karpuzoglu E (2012) Gender differences in cancer susceptibility: an inadequately addressed issue. Front Genet 3:268. https://doi.org/10.3389/fgene.2012.00268
Fan T, Li C, He J (2021) Prognostic value of immune-related genes and comparative analysis of immune cell infiltration in lung adenocarcinoma: sex differences. Biol Sex Differ 12(1):64. https://doi.org/10.1186/s13293-021-00406-y
Fang H, Disteche CM, Berletch JB (2019) X inactivation and escape: epigenetic and structural features. Front Cell Dev Biol 7:219. https://doi.org/10.3389/fcell.2019.00219
Furlan G, Gutierrez Hernandez N, Huret C, Galupa R, van Bemmel JG, Romito A, Heard E, Morey C, Rougeulle C (2018) The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol Cell 70(3):462-472.e8. https://doi.org/10.1016/j.molcel.2018.03.024
Gong D, Chen M, Wang Y, Shi J, Hou Y (2022) Role of ferroptosis on tumor progression and immunotherapy. Cell Death Discov 8(1):1. https://doi.org/10.1038/s41420-022-01218-8
Guo Y, Qu Z, Li D, Bai F, **ng J, Ding Q, Zhou J, Yao L, Xu Q (2021) Identification of a prognostic ferroptosis-related lncRNA signature in the tumor microenvironment of lung adenocarcinoma. Cell Death Discov 7(1):1. https://doi.org/10.1038/s41420-021-00576-z
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291):1071–1076. https://doi.org/10.1038/nature08975
Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12(1):31–46. https://doi.org/10.1158/2159-8290.CD-21-1059
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. https://doi.org/10.1016/j.cell.2011.02.013
Hartman RJG, Huisman SE, den Ruijter HM (2018) Sex differences in cardiovascular epigenetics—a systematic review. Biol Sex Differ 9(1):19. https://doi.org/10.1186/s13293-018-0180-z
He D, Li L, Zhu G, Liang L, Guan Z, Chang L, Chen Y, Yeh S, Chang C (2014) ASC-J9 suppresses renal cell carcinoma progression by targeting an androgen receptor-dependent HIF2α/VEGF signaling pathway. Cancer Res 74(16):4420–4430. https://doi.org/10.1158/0008-5472.CAN-13-2681
Henderson J (2005) Ernest starling and ‘hormones’: an historical commentary. J Endocrinol 184(1):5–10. https://doi.org/10.1677/joe.1.06000
Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, Thomas CC, Islami F, Weir HK, Lewis DR, Sherman RL, Wu M, Benard VB, Richardson LC, Jemal A, Cronin K, Kohler BA (2020) Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer 126(10):2225–2249. https://doi.org/10.1002/cncr.32802
Hiam-Galvez KJ, Allen BM, Spitzer MH (2021) Systemic immunity in cancer. Nat Rev Cancer 21(6):6. https://doi.org/10.1038/s41568-021-00347-z
Huang R, Zhu L, Zhang Y (2020) XIST lost induces ovarian cancer stem cells to acquire taxol resistance via a KMT2C-dependent way. Cancer Cell Int 20(1):436. https://doi.org/10.1186/s12935-020-01500-8
Isaev K, Jiang L, Wu S, Lee CA, Watters V, Fort V, Tsai R, Coutinho FJ, Hussein SMI, Zhang J, Wu J, Dirks PB, Schramek D, Reimand J (2021) Pan-cancer analysis of non-coding transcripts reveals the prognostic onco-lncRNA HOXA10-AS in gliomas. Cell Rep 37(3):109873. https://doi.org/10.1016/j.celrep.2021.109873
Islami F, Ward EM, Sung H, Cronin KA, Tangka FKL, Sherman RL, Zhao J, Anderson RN, Henley SJ, Yabroff KR, Jemal A, Benard VB (2021) Annual report to the nation on the status of cancer, part 1: national cancer statistics. JNCI J Natl Cancer Inst 113(12):1648–1669. https://doi.org/10.1093/jnci/djab131
Issler O, van der Zee YY, Ramakrishnan A, Wang J, Tan C, Loh Y-HE, Purushothaman I, Walker DM, Lorsch ZS, Hamilton PJ, Peña CJ, Flaherty E, Hartley BJ, Torres-Berrío A, Parise EM, Kronman H, Duffy JE, Estill MS, Calipari ES et al (2020) Sex-specific role for the long non-coding RNA LINC00473 in depression. Neuron 106(6):912-926.e5. https://doi.org/10.1016/j.neuron.2020.03.023
Jacobson EC, Pandya-Jones A, Plath K (2022) A lifelong duty: how **st maintains the inactive X chromosome. Curr Opin Genet Dev 75:101927. https://doi.org/10.1016/j.gde.2022.101927
Jiang S, Cheng S-J, Ren L-C, Wang Q, Kang Y-J, Ding Y, Hou M, Yang X-X, Lin Y, Liang N, Gao G (2019) An expanded landscape of human long noncoding RNA. Nucleic Acids Res 47(15):7842–7856. https://doi.org/10.1093/nar/gkz621
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F (2017) Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12(4):828–863. https://doi.org/10.1038/nprot.2017.016
Jusic A, Salgado-Somoza A, Paes AB, Stefanizzi FM, Martínez-Alarcón N, Pinet F, Martelli F, Devaux Y, Robinson EL, Novella S (2020) Approaching sex differences in cardiovascular non-coding RNA research. Int J Mol Sci 21(14):4890. https://doi.org/10.3390/ijms21144890
Karner H, Webb C-H, Carmona S, Liu Y, Lin B, Erhard M, Chan D, Baldi P, Spitale RC, Sun S (2020) Functional conservation of LncRNA JPX despite sequence and structural divergence. J Mol Biol 432(2):283–300. https://doi.org/10.1016/j.jmb.2019.09.002
Kirchgessner CU, Warren ST, Willard HF (1995) X inactivation of the FMR1 fragile X mental retardation gene. J Med Genet 32(12):925–929
Klein SL, Flanagan KL (2016) Sex differences in immune responses. Nat Rev Immunol 16(10):626–638. https://doi.org/10.1038/nri.2016.90
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172(3):393–407. https://doi.org/10.1016/j.cell.2018.01.011
Kumar S, Prajapati KS, Singh AK, Kushwaha PP, Shuaib M, Gupta S (2021) Long non-coding RNA regulating androgen receptor signaling in breast and prostate cancer. Cancer Lett 504:15–22. https://doi.org/10.1016/j.canlet.2020.11.039
Laskar RS, Li P, Ecsedi S, Abedi-Ardekani B, Durand G, Robinot N, Hubert J-N, Janout V, Zaridze D, Mukeria A, Mates D, Holcatova I, Foretova L, Swiatkowska B, Dzamic Z, Milosavljevic S, Olaso R, Boland A, Deleuze J-F et al (2021) Sexual dimorphism in cancer: Insights from transcriptional signatures in kidney tissue and renal cell carcinoma. Hum Mol Genet 30(5):343–355. https://doi.org/10.1093/hmg/ddab031
Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res 75(18):3728–3737. https://doi.org/10.1158/0008-5472.CAN-15-0273
Li M, Chai H-F, Peng F, Meng Y-T, Zhang L-Z, Zhang L, Zou H, Liang Q-L, Li M-M, Mao K-G, Sun D-X, Tong M-Y, Deng Z-Q, Hou Z-J, Zhao Y, Li J, Wang X-C, Lv S-S, Zhang Q-Q et al (2018) Estrogen receptor β upregulated by lncRNA-H19 to promote cancer stem-like properties in papillary thyroid carcinoma. Cell Death Dis 9(11):1120. https://doi.org/10.1038/s41419-018-1077-9
Li CH, Prokopec SD, Sun RX, Yousif F, Schmitz N, PCAWG Tumour Subtypes and Clinical Translation, Al-Shahrour F, Atwal G, Bailey PJ, Biankin AV, Boutros PC, Campbell PJ, Chang DK, Cooke SL, Deshpande V, Faltas BM, Faquin WC, Garraway L,Getz G et al (2020) Sex differences in oncogenic mutational processes. Nat Commun 11(1):4330. https://doi.org/10.1038/s41467-020-17359-2
Li J, Ming Z, Yang L, Wang T, Liu G, Ma Q (2022) Long noncoding RNA XIST: mechanisms for X chromosome inactivation, roles in sex-biased diseases, and therapeutic opportunities. Genes Dis 9(6):1478–1492. https://doi.org/10.1016/j.gendis.2022.04.007
Li J, Lan Z, Liao W, Horner JW, Xu X, Liu J, Yoshihama Y, Jiang S, Shim HS, Slotnik M, LaBella KA, Wu C-J, Dunner K, Hsu W-H, Lee R, Khanduri I, Terranova C, Akdemir K, Chakravarti D et al (2023a) Histone demethylase KDM5D upregulation drives sex differences in colon cancer. Nature 619(7970):632–639. https://doi.org/10.1038/s41586-023-06254-7
Li Y, Wei C, Huang C, Ling Q, Zhang L, Huang S, Liao N, Liang W, Cheng J, Wang F, Mo L, Mo Z, Li L (2023b) Long noncoding RNA as a potential diagnostic tool for prostate cancer: a systematic review and meta-analysis. Biomarkers 28(1):1–10. https://doi.org/10.1080/1354750X.2022.2142293
Liang J, ** W, Xu H (2022) An efficient five-lncRNA signature for lung adenocarcinoma prognosis, with AL606489.1 showing sexual dimorphism. Front Genet 13:1052092. https://doi.org/10.3389/fgene.2022.1052092
Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355(6320):aah7111. https://doi.org/10.1126/science.aah7111
Liu Y, Cao Z, Wang Y, Guo Y, Xu P, Yuan P, Liu Z, He Y, Wei W (2018) Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol. https://doi.org/10.1038/nbt.4283
Liu S, Lai W, Shi Y, Liu N, Ouyang L, Zhang Z, Chen L, Wang X, Qian B, **ao D, Yan Q, Cao Y, Liu S, Tao Y (2020) Annotation and cluster analysis of long noncoding RNA linked to male sex and estrogen in cancers. Npj Precis Oncol 4(1):5. https://doi.org/10.1038/s41698-020-0110-5
Lu S, Shan N, Chen X, Peng F, Wang Y, Long H (2021) A novel immune-related long non-coding RNAs risk model for prognosis assessment of lung adenocarcinoma. Aging 13(23):25550–25563. https://doi.org/10.18632/aging.203772
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:4773. https://doi.org/10.1038/190372a0
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15(suppl 1):R17–R29. https://doi.org/10.1093/hmg/ddl046
Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen L-L, Chen R, Dean C, Dinger ME, Fitzgerald KA, Gingeras TR, Guttman M, Hirose T, Huarte M, Johnson R, Kanduri C, Kapranov P, Lawrence JB, Lee JT et al (2023) Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. https://doi.org/10.1038/s41580-022-00566-8
Nair L, Chung H, Basu U (2020) Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol 21(3):123–136. https://doi.org/10.1038/s41580-019-0209-0
Patrat C, Ouimette J-F, Rougeulle C (2020) X chromosome inactivation in human development. Development 147(1):dev183095. https://doi.org/10.1242/dev.183095
Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42(1):1. https://doi.org/10.1146/annurev.genet.42.110807.091711
Payer B, Lee JT, Namekawa SH (2011) X-inactivation and X-reactivation: epigenetic hallmarks of mammalian reproduction and pluripotent stem cells. Hum Genet 130(2):265–280. https://doi.org/10.1007/s00439-011-1024-7
Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA (2014) Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat Rev Immunol 14(3):181–194. https://doi.org/10.1038/nri3623
Prolo LM, Li A, Owen SF, Parker JJ, Foshay K, Nitta RT, Morgens DW, Bolin S, Wilson CM, Vega LJCM, Luo EJ, Nwagbo G, Waziri A, Li G, Reimer RJ, Bassik MC, Grant GA (2019) Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. Sci Rep 9(1):14020. https://doi.org/10.1038/s41598-019-50160-w
Pyfrom S, Paneru B, Knox JJ, Cancro MP, Posso S, Buckner JH, Anguera MC (2021) The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc Natl Acad Sci USA 118(24):e2024624118. https://doi.org/10.1073/pnas.2024624118
Qi P, Zhou X, Du X (2016) Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 15(1):39. https://doi.org/10.1186/s12943-016-0524-4
Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17(1):1. https://doi.org/10.1038/nrg.2015.10
Ramilowski JA, Yip CW, Agrawal S, Chang J-C, Ciani Y, Kulakovskiy IV, Mendez M, Ooi JLC, Ouyang JF, Parkinson N, Petri A, Roos L, Severin J, Yasuzawa K, Abugessaisa I, Akalin A, Antonov IV, Arner E, Bonetti A et al (2020) Functional annotation of human long noncoding RNAs via molecular phenoty**. Genome Res 30(7):1060–1072. https://doi.org/10.1101/gr.254219.119
Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW, Jazin E (2010) Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genom 11(1):614. https://doi.org/10.1186/1471-2164-11-614
Richart L, Picod-Chedotel M-L, Wassef M, Macario M, Aflaki S, Salvador MA, Héry T, Dauphin A, Wicinski J, Chevrier V, Pastor S, Guittard G, Le Cam S, Kamhawi H, Castellano R, Guasch G, Charafe-Jauffret E, Heard E, Margueron R, Ginestier C (2022) XIST loss impairs mammary stem cell differentiation and increases tumorigenicity through mediator hyperactivation. Cell 185(12):2164-2183.e25. https://doi.org/10.1016/j.cell.2022.04.034
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81(1):145–166. https://doi.org/10.1146/annurev-biochem-051410-092902
Roberts AL, Morea A, Amar A, Zito A, El-Sayed Moustafa JS, Tomlinson M, Bowyer RC, Zhang X, Christiansen C, Costeira R, Steves CJ, Mangino M, Bell JT, Wong CC, Vyse TJ, Small KS (2022) Age acquired skewed X chromosome inactivation is associated with adverse health outcomes in humans. Elife 11:e78263. https://doi.org/10.7554/eLife.78263
Rosspopoff O, Cazottes E, Huret C, Loda A, Collier AJ, Casanova M, Rugg-Gunn PJ, Heard E, Ouimette J-F, Rougeulle C (2023) Species-specific regulation of XIST by the JPX/FTX orthologs. Nucleic Acids Res 51(5):2177–2194. https://doi.org/10.1093/nar/gkad029
Rubin JB, Lagas JS, Broestl L, Sponagel J, Rockwell N, Rhee G, Rosen SF, Chen S, Klein RS, Imoukhuede P, Luo J (2020) Sex differences in cancer mechanisms. Biol Sex Differ 11(1):17. https://doi.org/10.1186/s13293-020-00291-x
Sadagopan A, Nasim IT, Li J, Achom M, Zhang C-Z, Viswanathan SR (2022) Somatic XIST activation and features of X chromosome inactivation in male human cancers. Cell Syst 13(11):932-944.e5. https://doi.org/10.1016/j.cels.2022.10.002
Sandovici I, Fernandez-Twinn DS, Hufnagel A, Constância M, Ozanne SE (2022) Sex differences in the intergenerational inheritance of metabolic traits. Nat Metabol 4(5):5. https://doi.org/10.1038/s42255-022-00570-4
Schafer JM, **ao T, Kwon H, Collier K, Chang Y, Abdel-Hafiz H, Bolyard C, Chung D, Yang Y, Sundi D, Ma Q, Theodorescu D, Li X, Li Z (2022) Sex-biased adaptive immune regulation in cancer development and therapy. iScience 25(8):104717. https://doi.org/10.1016/j.isci.2022.104717
Schmidt K, Carroll JS, Yee E, Thomas DD, Wert-Lamas L, Neier SC, Sheynkman G, Ritz J, Novina CD (2019) The lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in melanoma and inhibits expression of tumor suppressor p21. Cell Rep 27(8):2493-2507.e4. https://doi.org/10.1016/j.celrep.2019.04.101
Simchovitz-Gesher A, Soreq H (2020) Pharmaceutical implications of sex-related RNA divergence in psychiatric disorders. Trends Pharmacol Sci 41(11):840–850. https://doi.org/10.1016/j.tips.2020.09.003
Sohail SK, Sarfraz R, Imran M, Kamran M, Qamar S (2020) Estrogen and progesterone receptor expression in breast carcinoma and its association with clinicopathological variables among the Pakistani population. Cureus 12(8):e9751. https://doi.org/10.7759/cureus.9751
Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT (2013) Jpx RNA activates **st by evicting CTCF. Cell 153(7):1537–1551. https://doi.org/10.1016/j.cell.2013.05.028
Syrett CM, Paneru B, Sandoval-Heglund D, Wang J, Banerjee S, Sindhava V, Behrens EM, Atchison M, Anguera MC (2019) Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4(7):e126751. https://doi.org/10.1172/jci.insight.126751
Taheri M, Safarzadeh A, Hussen BM, Ghafouri-Fard S, Baniahmad A (2022) LncRNA/miRNA/mRNA network introduces novel biomarkers in prostate cancer. Cells 11(23):3776. https://doi.org/10.3390/cells11233776
Tao S, He H, Chen Q (2015) Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J Transl Med. https://doi.org/10.1186/s12967-015-0489-x
The Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474(7353):609–615. https://doi.org/10.1038/nature10166
Tietze L, Kessler SM (2020) The good, the bad, the question-H19 in hepatocellular carcinoma. Cancers 12(5):1261. https://doi.org/10.3390/cancers12051261
Viggiano E, Ergoli M, Picillo E, Politano L (2016) Determining the role of skewed X-chromosome inactivation in develo** muscle symptoms in carriers of Duchenne muscular dystrophy. Hum Genet 135(7):685–698. https://doi.org/10.1007/s00439-016-1666-6
Wang R, Sun Y, Li L, Niu Y, Lin W, Lin C, Antonarakis ES, Luo J, Yeh S, Chang C (2017) Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9® to suppress enzalutamide-resistant prostate cancer progression. Eur Urol 72(5):835–844. https://doi.org/10.1016/j.eururo.2017.04.005
Wutz A (2011) Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet 12(8):8. https://doi.org/10.1038/nrg3035
**ng F, Liu Y, Wu S-Y, Wu K, Sharma S, Mo Y-Y, Feng J, Sanders S, ** G, Singh R, Vidi P-A, Tyagi A, Chan MD, Ruiz J, Debinski W, Pasche BC, Lo H-W, Metheny-Barlow LJ, D’Agostino RB, Watabe K (2018) Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal microRNA to promote brain metastasis. Cancer Res 78(15):4316–4330. https://doi.org/10.1158/0008-5472.CAN-18-1102
Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K, Liu X, Xu T, Sun L, Qin J, He B, Pan Y, Sun H, Wang S (2019) LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer 18(1):135. https://doi.org/10.1186/s12943-019-1063-6
Xue X, Yang YA, Zhang A, Fong K-W, Kim J, Song B, Li S, Zhao JC, Yu J (2016) LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 35(21):2746–2755. https://doi.org/10.1038/onc.2015.340
Yager JD, Davidson NE (2006) Estrogen carcinogenesis in breast cancer. N Engl J Med 354(3):270–282. https://doi.org/10.1056/NEJMra050776
Yao R-W, Wang Y, Chen L-L (2019) Cellular functions of long noncoding RNAs. Nat Cell Biol 21(5):5. https://doi.org/10.1038/s41556-019-0311-8
Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT (2013) **st RNA is a potent suppressor of hematologic cancer in mice. Cell 152:4. https://doi.org/10.1016/j.cell.2013.01.034
Yip CW, Hon C-C, Yasuzawa K, Sivaraman DM, Ramilowski JA, Shibayama Y, Agrawal S, Prabhu AV, Parr C, Severin J, Lan YJ, Dostie J, Petri A, Nishiyori-Sueki H, Tagami M, Itoh M, López-Redondo F, Kouno T, Chang J-C et al (2022) Antisense-oligonucleotide-mediated perturbation of long non-coding RNA reveals functional features in stem cells and across cell types. Cell Rep 41(13):111893. https://doi.org/10.1016/j.celrep.2022.111893
Yu W, Ding J, He M, Chen Y, Wang R, Han Z, **ng EZ, Zhang C, Yeh S (2019) Estrogen receptor β promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 38(8):1225–1238. https://doi.org/10.1038/s41388-018-0463-1
Yu B, Qi Y, Li R, Shi Q, Satpathy AT, Chang HY (2021) B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 184(7):1790-1803.e17. https://doi.org/10.1016/j.cell.2021.02.015
Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng J, Lin W, Huang Q, Li G, Zheng J, Chang C (2017) LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ 24(9):1502–1517. https://doi.org/10.1038/cdd.2017.74
Zhang J, Fan D, Jian Z, Chen GG, Lai PBS (2015) Cancer specific long noncoding RNAs show differential expression patterns and competing endogenous RNA potential in hepatocellular carcinoma. PLoS One 10(10):e0141042. https://doi.org/10.1371/journal.pone.0141042
Zhang Y, Pitchiaya S, Cieślik M, Niknafs YS, Tien JC-Y, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla SK, Jiang X, Wang L, Liu T-Y, Uhl M, Gawronski AR, Qiao Y, **ao L, Dhanasekaran SM, Juckette KM et al (2018) Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 50(6):814–824. https://doi.org/10.1038/s41588-018-0120-1
Acknowledgements
The authors would like to thank members of the Sun Lab and the Kong Lab for discussion and feedback of this work. The authors would also like to thank the three anonymous reviewers for helpful comments to improve the manuscript.
Funding
This publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award No. P30CA062203 and the UC Irvine Comprehensive Cancer Center using UCI Anti-Cancer Challenge funds. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Chao Family Comprehensive Cancer Center. This work was also funded in part by the National Institutes of Health through National Cancer Institute Award R01CA244360 (M.K.) and the National Institute of General Medical Sciences Award R01GM141424 (S.S.).
Author information
Authors and Affiliations
Contributions
IN, MDA-L, YY, MK, and SS drafted and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Naciri, I., Andrade-Ludena, M.D., Yang, Y. et al. An emerging link between lncRNAs and cancer sex dimorphism. Hum. Genet. (2023). https://doi.org/10.1007/s00439-023-02620-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00439-023-02620-7