Abstract
The Oryza genus, containing Oryza sativa L., is quintessential to sustain global food security. This genus has a lot of sophisticated molecular mechanisms to cope with environmental stress, particularly during vulnerable stages like flowering. Recent studies have found key involvements and genetic modifications that increase resilience to stress, including exogenous application of melatonin, allantoin, and trehalose as well as OsSAPK3 and OsAAI1 in the genetic realm. Due to climate change and anthropogenic reasons, there is a rise in sea level which raises a concern of salinity stress. It is tackled through osmotic adjustment and ion homeostasis, mediated by genes like P5CS, P5CR, GSH1, GSH2, and SPS, and ion transporters like NHX, NKT, and SKC, respectively. Oxidative damage is reduced by a complex action of antioxidants, scavenging RONS. A complex action of genes mediates cold stress with studies highlighting the roles of OsWRKY71, microRNA2871b, OsDOF1, and OsICE1. There is a need to research the mechanism of action of proteins like OsRbohA in ROS control and the action of regulatory genes in stress response. This is highly relevant due to the changing climate which will raise a lot of environmental changes that will adversely affect production and global food security if certain countermeasures are not taken. Overall, this study aims to unravel the molecular intricacies of ROS and RNS signaling networks in Oryza plants under stress conditions, with the ultimate goal of informing strategies for enhancing stress tolerance and crop performance in this important agricultural genus.
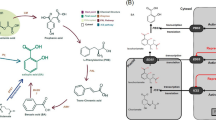
Graphical abstract


(Adopted from Kapoor et al. 2019; Nieves-Cordones et al. 2019; Romero-Puertas et al. 2019; Mukherjee et al. 2020; Enciso-Martínez et al. 2024)

(Adopted from Kapoor et al. 2019; Nieves-Cordones et al. 2019; Romero-Puertas et al. 2019; Mukherjee et al. 2020; Enciso-Martínez et al. 2024)

(Adopted from Kapoor et al. 2019; Nieves-Cordones et al. 2019; Romero-Puertas et al. 2019; Mukherjee et al. 2020; Enciso-Martínez et al. 2024)

(Adopted from Kapoor et al. 2019; Nieves-Cordones et al. 2019; Romero-Puertas et al. 2019; Mukherjee et al. 2020; Enciso-Martínez et al. 2024)
Similar content being viewed by others
Data availability
Data to support the findings of this study is available on reasonable request from the corresponding author.
Abbreviations
- ROS:
-
Reactive oxygen species
- RNS:
-
Reactive nitrogen species
- PCD:
-
Programmed cell death
- •OH:
-
Hydroxyl radicals
- H2O2 :
-
Hydrogen peroxide
- •NO:
-
Nitric monoxide
- ER:
-
Endoplasmic reticulum
- O2 :
-
Oxygen
- NO+ :
-
Nitrosonium
- ONOO− :
-
Peroxynitrite
- ANOVA:
-
Analysis of variance
- POD:
-
Peroxidase
- APX:
-
Ascorbate peroxidase
- GR:
-
Glutathione reductase
- CAT:
-
Catalase
- PEG:
-
Polyethylene glycol
- SL:
-
Sister line
- IPCC:
-
Intergovernmental panel on climate change
- IAA:
-
Indole-3-acetic acid
- LRR:
-
Leucine-rich repeat
- LTS:
-
Low-temperature stress
- TBARS:
-
Thiobarbituric acid reactive substances
- MDA:
-
Malondialdehyde
- DXWR:
-
Dongxiang wild rice
- BiFC:
-
Bimolecular fluorescence complementation
- BD:
-
Binding domain
- AD:
-
Activation domain
- RCPs:
-
Representative concentration pathways
- HS:
-
Heat stress
- DS:
-
Drought stress
- CK:
-
Water treatment
- GO:
-
Gene ontology
- BR:
-
Brassinosteroids
- HSPs:
-
Heat shock proteins
- ERF:
-
Ethylene-responsive factor
- DREB:
-
Dehydration-responsive element-binding
- MYB:
-
Myeloblastosis
- SOD:
-
Superoxide dismutase
- aCTK:
-
Active cytokinins
- aGA:
-
Active gibberellins
- ABA:
-
Abscisic acid
- AVG:
-
Aminoethoxyvinylglycine
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- BA:
-
Boric acid
- SB:
-
Sodium borate
- QTL:
-
Quantitative trait loci
- GST:
-
Glutathione-S-transferase
- AOS:
-
Antioxidative systems
- XOR:
-
Xanthine-oxidoreductase
- NiR:
-
Nitrite reductase
- GSH:
-
Glutathione
- WGCNA:
-
Weighted correlation network analysis
- AOS:
-
Antioxidant system
- ETC:
-
Electron transport chain
- PRRs:
-
Pattern recognition receptors
- DAMPs:
-
Damage-associated molecular patterns
- SOS:
-
Salt overly sensitive
References
Aamir M, Singh VK, Meena M, Upadhyay RS, Gupta VK, Singh S (2017) Structural and functional insights into WRKY3 and WRKY4 transcription factors to unravel the WRKY–DNA (W-box) complex interaction in tomato (Solanum lycopersicum) (L) A computational approach. Front Plant Sci 8:819. https://doi.org/10.3389/fpls.2017.00819
Abisha R, Krishnani KK, Sukhdhane K, Verma AK, Brahmane M, Chadha NK (2022) Sustainable development of climate-resilient aquaculture and culture-based fisheries through adaptation of abiotic stresses: A review. J Water Clim Chang 13(7):2671–2689. https://doi.org/10.2166/wcc.2022.045
Agati G, Azzarello E, Pollastri S, Tattini M (2012) Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci 196:67–76. https://doi.org/10.1016/j.plantsci.2012.07.014
Ahmar S, Gruszka D (2022) In-silico study of brassinosteroid signaling genes in rice provides insight into mechanisms which regulate their expression. Front Genet 13:953458. https://doi.org/10.3389/fgene.2022.953458
Ai P, Xue J, Shi Z, Liu Y, Li Z, Li T, Zhao W, Khan MA, Kang D, Wang K, Wang Z (2023) Genome-wide characterization and expression analysis of MYB transcription factors in Chrysanthemum nankingense. BMC Plant Biol 23(1):140. https://doi.org/10.1186/s12870-023-04137-7
Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, signal transduction. Ann Rev Plant Biol 55(1):373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Astier J, Gross I, Durner J (2018) Nitric oxide production in plants: An update. J Exp Bot 69(14):3401–3411. https://doi.org/10.1093/jxb/erx420
Azameti MK, Dauda WP, Panzade KP, Vishwakarma H (2021) Identification and characterization of genes responsive to drought and heat stress in rice (Oryza sativa L). Vegetos 34(2):309–317. https://doi.org/10.1007/s42535-021-00198-x
Bandumula N (2018) Rice production in Asia: Key to global food security. Proc Natl Acad Sci India Sec B Biol Sci 88(4):1323–1328. https://doi.org/10.1007/s40011-017-0867-7
Barman D, Kumar R, Ghimire OP, Ramesh R, Gupta S, Nagar S, Pal M, Dalal M, Chinnusamy V, Arora A (2024) Melatonin induces acclimation to heat stress and pollen viability by enhancing antioxidative defense in rice (Oryza Sativa L). Environ Exp Bot 13:105693. https://doi.org/10.1016/j.envexpbot.2024.105693
Berger A, Boscari A, Horta Araújo N, Maucourt M, Hanchi M, Bernillon S, Rolin D, Puppo A, Brouquisse R (2020) Plant nitrate reductases regulate nitric oxide production and nitrogen-fixing metabolism during the Medicago truncatula–Sinorhizobium meliloti symbiosis. Front Plant Sci 11:1313. https://doi.org/10.3389/fpls.2020.01313
Bhupenchandra I, Chongtham SK, Devi AG, Dutta P, Lamalakshmi E, Mohanty S, Choudhary AK, Das A, Sarika K, Kumar S, Yumnam S, Sagolsem D, Anand YR, Bhutia DD, Victoria M, Vinodh S, Tania C, Sharma AD, Deb L, Sahoo MR, Seth CS, Swapnil P, Meena M (2024) Harnessing weedy rice as functional food and source of novel traits for crop improvement. Plant Cell Environ. https://doi.org/10.1111/pce.14868
Burke R, Schwarze J, Sherwood OL, Jnaid Y, McCabe PF, Kacprzyk J (2020) Stressed to death: The role of transcription factors in plant programmed cell death induced by abiotic and biotic stimuli. Front Plant Sci 11:1235. https://doi.org/10.3389/fpls.2020.01235
Calderón-Páez SE, Cueto-Niño YA, Sánchez-Reinoso AD, Garces-Varon G, Chávez-Arias CC, Restrepo-Díaz H (2021) Foliar boron compounds applications mitigate heat stress caused by high daytime temperatures in rice (Oryza sativa L) Boron mitigates heat stress in rice. J Plant Nutr 44(17):2514–2527. https://doi.org/10.1080/01904167.2021.1921202
Calingacion M, Laborte A, Nelson A, Resurreccion A, Concepcion JC, Daygon VD, Mumm R, Reinke R, Dipti S, Bassinello PZ, Manful J, Sophany S, Lara KC, Bao J, **e L, Loaiza K, El-hissewy A, Gayin J, Sharma N, Fitzgerald M (2014) Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS ONE 9(1):e85106. https://doi.org/10.1371/journal.pone.0085106
Castro JLS, Lima-Melo Y, Carvalho FEL, Feitosa AGS, Lima Neto MC, Caverzan A, Margis-Pinheiro M, Silveira JAG (2018) Ascorbic acid toxicity is related to oxidative stress and enhanced by high light and knockdown of chloroplast ascorbate peroxidases in rice plants. Theor Exp Plant Physiol 30(1):41–55. https://doi.org/10.1007/s40626-018-0100-y
Chae HB, Kim MG, Kang CH, Park JH, Lee ES, Lee SU, Chi YH, Paeng SK, Bae SB, Wi SD, Yun BW, Kim WY, Yun DJ, Mackey D, Lee SY (2021) Redox sensor QSOX1 regulates plant immunity by targeting GSNOR to modulate ROS generation. Mol Plant 14(8):1312–1327. https://doi.org/10.1016/j.molp.2021.05.004
Chandran H, Meena M, Swapnil P (2021) Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustainability 13(19):10986. https://doi.org/10.3390/su131910986
Chen W, Zhao X (2023) Understanding global rice trade flows: Network evolution and implications. Foods 12(17):298. https://doi.org/10.3390/foods12173298
Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, Zuo J (2009) The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19(12):1377–1387. https://doi.org/10.1038/cr.2009.117
Chen HC, Cheng WH, Hong CY, Chang YS, Chang MC (2018) The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 11(1):50. https://doi.org/10.1186/s12284-018-0244-z
Chen Y, Li R, Ge J, Liu J, Wang W, Xu M, Zhang R, Hussain S, Wei H, Dai Q (2021) Exogenous melatonin confers enhanced salinity tolerance in rice by blocking the ROS burst and improving Na+/K+ homeostasis. Environ Exp Bot 189:104530. https://doi.org/10.1016/j.envexpbot.2021.104530
Choudhary R, Rajput VD, Ghodake G, Ahmad F, Meena M, ul Rehman R, Prasad R, Sharma RK, Singh R, Seth CS, (2024) Comprehensive journey from past to present to future about seed priming with hydrogen peroxide and hydrogen sulfide in relation to drought, temperature, UV and ozone stresses - A review. Plant Soil 494(1–2):1–23. https://doi.org/10.1007/s11104-024-06499-9
Corpas FJ, Barroso JB (2014) Peroxynitrite (ONOO-) is endogenously produced in arabidopsis peroxisomes and is overproduced under cadmium stress. Ann Bot 113(1):87–96. https://doi.org/10.1093/aob/mct260
Corpas FJ, Del Río LA, Barroso JB (2008) Post-translational modifications mediated by reactive nitrogen species: Nitrosative stress responses or components of signal transduction pathways? Plant Signal Behav 3(5):301–303. https://doi.org/10.4161/psb.3.5.5277
Correa-Aragunde N, Foresi N, Lamattina L (2015) Nitric oxide is a ubiquitous signal for maintaining redox balance in plant cells: Regulation of ascorbate peroxidase as a case study. J Exp Bot 66(10):2913–2921. https://doi.org/10.1093/jxb/erv073
Dayton L (2014) Agribiotechnology: Blue-sky rice. Nature 514:7524. https://doi.org/10.1038/514S52a
De Freitas GM, Thomas J, Liyanage R, Lay JO, Basu S, Ramegowda V, Do Amaral MN, Benitez LC, Bolacel Braga EJ, Pereira A (2019) Cold tolerance response mechanisms revealed through comparative analysis of gene and protein expression in multiple rice genotypes. PLoS ONE 14(6):e0218019. https://doi.org/10.1371/journal.pone.0218019
DeFalco TA, Zipfel C (2021) Molecular mechanisms of early plant pattern-triggered immune signaling. Mol Cell 81(17):3449–3467. https://doi.org/10.1016/j.molcel.2021.07.029
Del Río LA (2015) ROS and RNS in plant physiology: An overview. J Exp Bot 66(10):2827–2837. https://doi.org/10.1093/jxb/erv099
Du H, Wu N, Fu J, Wang S, Li X, **ao J, **ong L (2012) A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63(18):6467–6480. https://doi.org/10.1093/jxb/ers300
Dumanović J, Nepovimova E, Natić M, Kuča K, Jaćević V (2021) The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front Plant Sci 11:552969. https://doi.org/10.3389/fpls.2020.552969
Gavgani HN, Fawaz R, Ehyaei N, Walls D, Pawlowski K, Fulgos R, Park S, Assar Z, Ghanbarpour A, Geiger JH (2022) A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb. J Biol Chem 298(1):101395. https://doi.org/10.1016/j.jbc.2021.101395
Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228(1):191–201. https://doi.org/10.1007/s00425-008-0729-x
Gibbs DJ, Conde JV, Berckhan S, Prasad G, Mendiondo GM, Holdsworth MJ (2015) Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol 169(1):23–31. https://doi.org/10.1104/pp.15.00338
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Gosal SS, Wani SH, Kang MS (2009) Biotechnology and drought tolerance. J Crop Improv 23(1):19–54. https://doi.org/10.1080/15427520802418251
Guo Z, Cai L, Liu C, Chen Z, Guan S, Ma W, Pan G (2022) Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L) at the reproductive stage. Sci Rep 12(1):6224. https://doi.org/10.1038/s41598-022-10420-8
Gupta KJ, Igamberdiev AU (2015) Compartmentalization of reactive oxygen species and nitric oxide production in plant cells An overview. In: Gupta KJ, Igamberdiev AU (eds) Reactive oxygen and nitrogen species signaling and communication in plants. Springer International Publishing, Switzerland
Gupta B, Shrestha J (2023) Abiotic stress adaptation and tolerance mechanisms in crop plants. Front Plant Sci 14:1278895. https://doi.org/10.3389/fpls.2023.1278895
Halliwell B (1987) Oxidative damage, lipid peroxidation and antioxidant protection in chloroplasts. Chem Phys Lipids 44(2–4):27–340. https://doi.org/10.1016/0009-3084(87)90056-9
Halliwell B, Gutteridge JMC (2007) Antioxidant defences: Endogenous and diet derived free radicals in biology and medicine. Oxford University Press, New York
Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, Matsugasako JI, Takeda Y (2008) Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol 49(6):925–933. https://doi.org/10.1093/pcp/pcn066
Hancock JT, Veal D (2021) Nitric oxide, other reactive signalling compounds, redox, and reductive stress. J Exp Bot 72(3):819–829. https://doi.org/10.1093/jxb/eraa331
Hoang TML, Williams B, Khanna H, Dale J, Mundree SG (2014) Physiological basis of salt stress tolerance in rice expressing the antiapoptotic gene SfIAP. Funct Plant Biol 41(11):1168–1177. https://doi.org/10.1071/FP13308
Hori K, Sun J (2022) Rice grain size and quality. Rice 15(1):33. https://doi.org/10.1186/s12284-022-00579-z
Hu X, Zhang A, Zhang J, Jiang M (2006) Abscisic acid is a key inducer of hydrogen peroxide production in leaves of maize plants exposed to water stress. Plant Cell Physiol 47(11):1484–1495. https://doi.org/10.1093/pcp/pcl014
Huang B, Chen J, Zhang J, Liu H, Tian M, Gu Y, Hu Y, Li Y, Liu Y, Huang Y (2011) Characterization of ADP-glucose pyrophosphorylase encoding genes in source and sink organs of maize. Plant Mol Biol Rep 29(3):563–572. https://doi.org/10.1007/s11105-010-0262-5
Huang L, Zhang M, Jia J, Zhao X, Huang X, Ji E, Ni L, Jiang M (2018) An atypical late embryogenesis abundant protein OsLEA5 plays a positive role in ABA-induced antioxidant defense in Oryza sativa L. Plant Cell Physiol 59(5):916–929. https://doi.org/10.1093/pcp/pcy035
Huang H, Ullah F, Zhou DX, Yi M, Zhao Y (2019) Mechanisms of ROS regulation of plant development and stress responses. Front Plant Sci 10:800. https://doi.org/10.3389/fpls.2019.00800
Huang M, Yin X, Chen J, Cao F (2021) Biochar application mitigates the effect of heat stress on rice (Oryza sativa L) by regulating the root-zone environment. Front Plant Sci 12:711725. https://doi.org/10.3389/fpls.2021.711725
Hussain S, Nanda S, Ashraf M, Siddiqui A, Masood S, Khaskheli M, Suleman M, Zhu L, Zhu C, Cao X, Kong Y, ** Q, Zhang J (2023) Interplay impact of exogenous application of abscisic acid (ABA) and brassinosteroids (BRs) in rice growth, physiology, and resistance under sodium chloride stress. Life 13(2):498. https://doi.org/10.3390/life13020498
Inupakutika MA, Sengupta S, Devireddy AR, Azad RK, Mittler R (2016) The evolution of reactive oxygen species metabolism. J Exp Bot 67(21):5933–5943. https://doi.org/10.1093/jxb/erw382
Jena KK, Nissila EAJ (2017) Genetic improvement of rice (Oryza sativa L). In: Campos H, Caligari PDS (eds) Genetic improvement of tropical crops. Springer International Publishing, Switzerland
Ježek P, Hlavatá L (2005) Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol 37(12):2478–2503. https://doi.org/10.1016/j.biocel.2005.05.013
Jia Y, Gu X, Chai J, Yao X, Cheng S, Liu L, He S, Peng Y, Zhang Q, Zhu Z (2023) Rice OsANN9 enhances drought tolerance through modulating ROS scavenging systems. Int J Mol Sci 24(24):17495. https://doi.org/10.3390/ijms242417495
Jiménez A, Sevilla F, Martí MC (2021) Reactive oxygen species homeostasis and circadian rhythms in plants. J Exp Bot 72(16):5825–5840. https://doi.org/10.1093/jxb/erab318
**g XQ, Li WQ, Zhou MR, Shi PT, Zhang R, Shalmani A, Muhammad I, Wang GF, Liu WT, Chen KM (2021) Rice carbohydrate-binding malectin-like protein, OsCBM1, contributes to drought-stress tolerance by participating in NADPH oxidase-mediated ROS production. Rice 14(1):100. https://doi.org/10.1186/s12284-021-00541-5
Kathirvel AK, Karuppasami KM, Dhashnamurthi V, Vellingiri G, Muthurajan R, Venugopal A, Kuppusamy A, Alagarsamy S (2024) Enhancement of grain yield in rice under combined drought and high-temperature stress conditions by maintaining photosynthesis through antioxidant enzyme activities by melatonin. Plant Physiol Rep 29:262–277. https://doi.org/10.1007/s40502-023-00773-1
Kaya H, Iwano M, Takeda S, Kanaoka MM, Kimura S, Abe M, Kuchitsu K (2015) Apoplastic ROS production upon pollination by RbohH and RbohJ in Arabidopsis. Plant Signal Behav 10(2):e989050. https://doi.org/10.4161/15592324.2014.989050
Khan M, Al Azzawi TNI, Imran M, Hussain A, Mun BG, Pande A, Yun BW (2021) Effects of lead (Pb)-induced oxidative stress on morphological and physio-biochemical properties of rice. Biocell 45(5):1413–1423. https://doi.org/10.32604/biocell.2021.015954
Khan M, Ali S, Al Azzawi TNI, SaqibS UF, Ayaz A, Zaman W (2023) The key roles of ROS and RNS as a signaling molecule in plant–microbe interactions. Antioxidants 12(2):268. https://doi.org/10.3390/antiox12020268
Kou S, Gu Q, Duan L, Liu G, Yuan P, Li H, Wu Z, Liu W, Huang P, Liu L (2022) Genome-wide bisulphite sequencing uncovered the contribution of DNA methylation to rice short-term drought memory formation. J Plant Growth Regul 41(7):2903–2917. https://doi.org/10.1007/s00344-021-10483-3
Kumar R, Swapnil P, Meena M, Selpair S, Yadav BG (2022a) Plant growth-promoting rhizobacteria (PGPR): Approaches to alleviate abiotic stresses for enhancement of growth and development of medicinal plants. Sustainability 14(23):15514. https://doi.org/10.3390/su142315514
Kumar S, Chandra R, Keswani C, Minkina·T, Mandzhieva S, Voloshina·M, Meena M, (2022) Trichoderma viride—mediated modulation of oxidative stress network in potato challenged with Alternaria solani. J Plant Growth Regul 42:1919–1936. https://doi.org/10.1007/s00344-022-10669-3
Kumar GA, Kumar S, Bhardwaj R, Meena M, Swapnil P, Seth CS, Yadav A (2024) Recent advancements in multifaceted roles of flavonoids in plant–rhizomicrobiome interactions. Front Plant Sci 14:1297706. https://doi.org/10.3389/fpls.2023.1297706
Laxa M, Liebthal M, Telman W, Chibani K, Dietz KJ (2019) The role of the plant antioxidant system in drought tolerance. Antioxidants 8(4):94. https://doi.org/10.3390/antiox8040094
Lee S, Sakuraba Y, Lee T, Kim K, An G, Lee HY, Paek N (2015) Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J Integr Plant Biol 57(6):562–576. https://doi.org/10.1111/jipb.12276
Li J, Khatab AA, Hu L, Zhao L, Yang J, Wang L, ** identifies new candidate genes for cold stress and chilling acclimation at seedling stage in rice (Oryza sativa L). Int J Mol Sci 23(21):13208. https://doi.org/10.3390/ijms232113208
Li B, Cai H, Liu K, An B, Wang R, Yang F, Zeng C, Jiao C, Xu Y (2023a) DNA methylation alterations and their association with high temperature tolerance in rice anthesis. J Plant Growth Regul 42(2):780–794. https://doi.org/10.1007/s00344-022-10586-5
Li Y, Han S, Sun X, Khan NU, Zhong Q, Zhang Z, Zhang H, Ming F, Li Z, Li J (2023b) Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice. J Integr Plant Biol 65(4):918–933. https://doi.org/10.1111/jipb.13414
Liu W, Ji S, Fang X, Wang Q, Li Z, Yao F, Hou L, Dai S (2013) Protein kinase LTRPK1 influences cold adaptation and microtubule stability in rice. J Plant Growth Regul 32(3):483–490. https://doi.org/10.1007/s00344-012-9314-4
Liu B, Zhao S, Tan F, Zhao H, Wang D, Si H, Chen Q (2017) Changes in ROS production and antioxidant capacity during tuber sprouting in potato. Food Chem 237:205–213. https://doi.org/10.1016/j.foodchem.2017.05.107
Liu J, Meng Q, **ang H, Shi F, Ma L, Li Y, Liu C, Liu Y, Su B (2021) Genome-wide analysis of Dof transcription factors and their response to cold stress in rice (Oryza sativa L). BMC Genom 22(1):800. https://doi.org/10.1186/s12864-021-08104-0
Liu L, Chao N, Yidilisi K, Kang X, Cao X (2022) Comprehensive analysis of the MYB transcription factor gene family in Morus alba. BMC Plant Biol 22(1):281. https://doi.org/10.1186/s12870-022-03626-5
Liu J, Meng Q, **ang H, Shi F, Ma L, Li Y, Yu L, Liu C, Liu Y, Su B (2023a) Transcription factor OsDOF1 enhances cold-stress tolerance potentially through interactions with OsICE1 and its target genes in rice (Oryza sativa L). Crop Sci 63(5):2963–2975. https://doi.org/10.1002/csc2.21073
Liu Q, Ma X, Li X, Zhang X, Zhou S, **ong L, Zhao Y, Zhou DX (2023b) Paternal DNA methylation is remodeled to maternal levels in rice zygote. Nat Commun 14(1):6571. https://doi.org/10.1038/s41467-023-42394-0
Long Q, Qiu S, Man J, Ren D, Xu N, Luo R (2023) OsAAI1 increases rice yield and drought tolerance dependent on ABA-mediated regulatory and ROS scavenging pathway. Rice 16(1):35. https://doi.org/10.1186/s12284-023-00650-3
Lopes Hornai EML, Aycan M, Mitsui T (2024) The promising B−type response regulator hst1 gene provides multiple high temperature and drought stress tolerance in rice. Int J Mol Sci 25(4):2385. https://doi.org/10.3390/ijms25042385
Luo C, Min W, Akhtar M, Lu X, Bai X, Zhang Y, Tian L, Li P (2022) Melatonin enhances drought tolerance in rice seedlings by modulating antioxidant systems, osmoregulation, corresponding gene expression. Int J Mol Sci 23(20):12075. https://doi.org/10.3390/ijms232012075
Lou D, Lu S, Chen Z, Lin Y, Yu D, Yang X (2023) Molecular characterization reveals that OsSAPK3 improves drought tolerance and grain yield in rice. BMC Plant Biol 23(1):53. https://doi.org/10.1186/s12870-023-04071-8
Lu H, Hu Y, Wang C, Liu W, Ma G, Han Q, Ma D (2019) Effects of high temperature and drought stress on the expression of gene encoding enzymes and the activity of key enzymes involved in starch biosynthesis in wheat grains. Front Plant Sci 10:1414. https://doi.org/10.3389/fpls.2019.01414
Lu R, Liu Z, Shao Y, Su J, Li X, Sun F, Zhang Y, Li S, Zhang Y, Cui J, Zhou Y, Shen W, Zhou T (2020) Nitric oxide enhances rice resistance to Rice Black-Streaked Dwarf Virus infection. Rice 13(1):24. https://doi.org/10.1186/s12284-020-00382-8
Lu S, Jia Z, Meng X, Chen Y, Wang S, Fu C, Yang L, Zhou R, Wang B, Cao Y (2022) Combined metabolomic and transcriptomic analysis reveals allantoin enhances drought tolerance in rice. Int J Mol Sci 23(22):14172. https://doi.org/10.3390/ijms232214172
Lu L, Sun Z, Wang R, Du Y, Zhang Z, Lan T, Song Y, Zeng R (2023) Integration of transcriptome and metabolome analyses reveals the role of OsSPL10 in rice defense against brown planthopper. Plant Cell Rep 42(12):2023–2038. https://doi.org/10.1007/s00299-023-03080-z
Ma Q, Hu Z, Mao Z, Mei Y, Feng S, Shi K (2022) The novel leucine-rich repeat receptor-like kinase MRK1 regulates resistance to multiple stresses in tomato. Hortic Res 9:088. https://doi.org/10.1093/hr/uhab088
Mabuchi K, Maki H, Itaya T, Suzuki T, Nomoto M, Sakaoka S, Morikami A, Higashiyama T, Tada Y, Busch W, Tsukagoshi H (2018) MYB30 links ROS signaling, root cell elongation, plant immune responses. Proc Natl Acad Sci 115(20):E4710–E4719. https://doi.org/10.1073/pnas.1804233115
Madison I, Gillan L, Peace J, Gabrieli F, Van Den Broeck L, Jones JL, Sozzani R (2023) Phosphate starvation: Response mechanisms and solutions. J Exp Bot 74(21):6417–6430. https://doi.org/10.1093/jxb/erad326
Mandal M, Sarkar M, Khan A, Biswas M, Masi A, Rakwal R, Agrawal GK, Srivastava A, Sarkar A (2022) Reactive oxygen species (ROS) and reactive nitrogen species (RNS) in plants–maintenance of structural individuality and functional blend. Adv Redox Res 5:100039. https://doi.org/10.1016/j.arres.2022.100039
Marro N, Caccia M, López-Ráez JA (2021) Are strigolactones a key in plant–parasitic nematodes interactions? An Intriguing Question. Plant Soil 462(1–2):591–601. https://doi.org/10.1007/s11104-021-04862-8
Meena M, Swapnil P (2019) Regulation of WRKY genes in plant defense with beneficial fungus Trichoderma: Current perspectives and future prospects. Arch Phytopathol Plant Protect 52(1–2):1–17. https://doi.org/10.1080/03235408.2019.1606490
Meena M, Yadav G, Sonigra P, Nagda A, Mehta T, Swapnil P, Harish MA (2022a) Role of elicitors to initiate the induction of systemic resistance in plants to biotic stress. Plant Stress 5:100103. https://doi.org/10.1016/j.stress.2022.100103
Meena M, Yadav G, Sonigra P, Nagda A, Mehta T, Zehra A, Swapnil P (2022b) Role of microbial bioagents as elicitors in plant defense regulation. In: Wani SH, Nataraj V, Singh GP (eds) Transcription factors for biotic stress tolerance in plants. Springer Nature, Switzerland
Mehta T, Meena M, Nagda A (2022) Bioactive compounds of Curvularia species as a source of various biological activities and biotechnological applications. Front Microbiol 13:1069095. https://doi.org/10.3389/fmicb.2022.1069095
Mhamdi A, Van Breusegem F (2018) Reactive oxygen species in plant development. Development 145(15):164376. https://doi.org/10.1242/dev.16437
Mishra D (2022) A big role for a microRNA in regulating cold tolerance and hormone signaling in rice. Plant Physiol 190(1):193–195. https://doi.org/10.1093/plphys/kiac292
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7(9):405–410. https://doi.org/10.1016/s1360-1385(02)02312-9
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9(10):490–498. https://doi.org/10.1016/j.tplants.2004.08.009
Mohanan A, Kodigudla A, Raman DR, Bakka K, Challabathula D (2023) Trehalose accumulation enhances drought tolerance by modulating photosynthesis and ROS-antioxidant balance in drought sensitive and tolerant rice cultivars. Physiol Mol Biol Plants 29(12):2035–2049. https://doi.org/10.1007/s12298-023-01404-7
Mohidem NA, Hashim N, Shamsudin R, Che Man H (2022) Rice for food security: Revisiting its production, diversity, rice milling process and nutrient content. Agriculture 12(6):741. https://doi.org/10.3390/agriculture12060741
Molnár Á, Rónavári A, Bélteky P, Szőllősi R, Valyon E, Oláh D, Rázga Z, Ördög A, Kónya Z, Kolbert Z (2020) ZnO nanoparticles induce cell wall remodeling and modify ROS/RNS signalling in roots of Brassica seedlings. Ecotoxicol Environ Saf 206:111158. https://doi.org/10.1016/j.ecoenv.2020.111158
Munné-Bosch S, Alegre L (2002) The function of tocopherols and tocotrienols in plants. Crit Rev Plant Sci 21(1):31–57. https://doi.org/10.1080/0735-260291044179
Munns R (2011) Plant adaptations to salt and water stress: Differences and commonalities. In: Turkan I (ed) Advances in botanical research. Academic Press, London
Munns R, James RA, Läuchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57(5):1025–1043. https://doi.org/10.1093/jxb/erj100
Mur LA, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJ, Hebelstrup KH, Gupta KJ (2013) Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 5:052. https://doi.org/10.1093/aobpla/pls052
Nazir F, Jahan B, Kumari S, Iqbal N, Albaqami M, Sofo A, Khan MI (2023) Brassinosteroid modulates ethylene synthesis and antioxidant metabolism to protect rice (Oryza sativa) against heat stress-induced inhibition of source-sink capacity and photosynthetic and growth attributes. J Plant Physiol 289:154096. https://doi.org/10.1016/j.jplph.2023.154096
Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8(1):68–82. https://doi.org/10.1016/j.molp.2014.12.007
Noctor G, De Paepe R, Foyer CH (2007) Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci 12(3):125–134. https://doi.org/10.1016/j.tplants.2007.01.005
Noctor G, Reichheld JP, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80:3–12. https://doi.org/10.1016/j.semcdb.2017.07.013
Pacher P, Beckman JS, Liaudet L (2007) Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87(1):315–424. https://doi.org/10.1152/physrev.00029.2006
Palma JM, Gupta DK, Corpas FJ (2019) Hydrogen peroxide and nitric oxide generation in plant cells: overview and queries. In: Gupta DK, Palma JM, Corpas FJ (eds) Nitric oxide and hydrogen peroxide signaling in higher plants. Springer International Publishing, Switzerland
Palma JM, Mateos Bernal RM, López-Jaramillo J, Rodríguez-Ruiz M, González-Gordo S, Lechuga-Sancho AM, Corpas FJ (2020) Plant catalases as NO and H2S targets. Redox Biol 34:101525. https://doi.org/10.1016/j.redox.2020.101525
Pantoja-Benavides AD, Garces-Varon G, Restrepo-Díaz H (2022) Foliar cytokinins or brassinosteroids applications influence the rice plant acclimatization to combined heat stress. Front Plant Sci 13:983276. https://doi.org/10.3389/fpls.2022.983276
Pastore D, Trono D, Laus MN, Di Fonzo N, Flagella Z (2007) Possible plant mitochondria involvement in cell adaptation to drought stress: A case study: durum wheat mitochondria. J Exp Bot 58(2):195–210. https://doi.org/10.1093/jxb/erl273
Patel CB, Singh VK, Singh AP, Meena M, Upadhyay RS (2019) Microbial genes involved in interaction with plants. In: Singh HB, Gupt VK, Jogaiah S (eds) New and future developments in microbial biotechnology and bioengineering. Elsevier, Amsterdam
Pfannschmidt T (2003) Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci 8(1):33–41. https://doi.org/10.1016/S1360-1385(02)00005-5
Prakash V, Singh VP, Tripathi DK, Sharma S, Corpas FJ (2021) Nitric oxide (NO) and salicylic acid (SA): A framework for their relationship in plant development under abiotic stress. Plant Biol 23(S1):39–49. https://doi.org/10.1111/plb.13246
Qi J, Wang J, Gong Z, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38:92–100. https://doi.org/10.1016/j.pbi.2017.04.022
Qi J, Song C, Wang B, Zhou J, Kangasjärvi J, Zhu J, Gong Z (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60(9):805–826. https://doi.org/10.1111/jipb.12654
Que Z, Lu Q, Shen C (2022) Chromosome-level genome assembly of Dongxiang wild rice (Oryza rufipogon) provides insights into resistance to disease and freezing. Front Genet 13:1029879. https://doi.org/10.3389/fgene.2022.1029879
Rajkumari N, Chowrasia S, Nishad J, Ganie SA, Mondal TK (2023) Metabolomics-mediated elucidation of rice responses to salt stress. Planta 258(6):111. https://doi.org/10.1007/s00425-023-04258-1
Rajput VD, Harish SRK, Verma KK, Sharma L, Quiroz-Figueroa FR, Meena M, Gour VS, Minkina T, Sushkova S, Mandzhieva S (2021) Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 10:267. https://doi.org/10.3390/biology10040267
Rathna Priya TS, Eliazer Nelson ARL, Ravichandran K, Antony U (2019) Nutritional and functional properties of coloured rice varieties of South India: A review. J Ethn Food 6(1):11. https://doi.org/10.1186/s42779-019-0017-3
Sabbioni G, Funck D, Forlani G (2021) Enzymology and regulation of δ1-pyrroline-5-carboxylate synthetase 2 from rice. Front Plant Sci 12:672702. https://doi.org/10.3389/fpls.2021.672702
Sakakibara H, Honda Y, Nakagawa S, Ashida H, Kanazawa K (2003) Simultaneous determination of all polyphenols in vegetables, fruits, teas. J Agric Food Chem 51(3):571–581. https://doi.org/10.1021/jf020926l
Sánchez-Bermúdez M, Del Pozo JC, Pernas M (2022) Effects of combined abiotic stresses related to climate change on root growth in crops. Front Plant Sci 13:918537. https://doi.org/10.3389/fpls.2022.918537
Sapna H, Ashwini N, Ramesh S, Nataraja KN (2020) Assessment of DNA methylation pattern under drought stress using methylation-sensitive randomly amplified polymorphism analysis in rice. Plant Genet Resour 18(4):222–230. https://doi.org/10.1017/S1479262120000234
Sarangthem K, Yumlembam S, Benazir S, Mikawlrawng K, Yendrembam R (2021) Current understanding of the mechanisms of heat stress tolerance in rice Oryza sativa L. J Exp Biol Agric Sci 9(3):S321–S329. https://doi.org/10.18006/2021.9(Spl-3-NRMCSSA_2021).S321.S329
Sarwar N, Atique-ur-Rehman AS, Hasanuzzaman M (2022) Modern techniques of rice crop production. Springer, Singapore
Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 2012:217037. https://doi.org/10.1155/2012/217037
Sharma S, Kaur C, Singla-Pareek SL, Sopory SK (2016) OsSRO1a interacts with RNA binding domain-containing protein (OsRBD1) and functions in abiotic stress tolerance in yeast. Front Plant Sci 7:62. https://doi.org/10.3389/fpls.2016.00062
Sharma V, Jahan K, Kumar P, Puri A, Sharma VK, Mishra A, Bharatam PV, Sharma D, Rishi V, Roy J (2023) Mechanistic insights into granule-bound starch synthase I (GBSSIL539P) allele in high amylose starch biosynthesis in wheat (Triticum aestivum L). Funct Integr Genom 23(1):20. https://doi.org/10.1007/s10142-022-00923-y
Simon I, Persky Z, Avital A, Harat H, Schroeder A, Shoseyov O (2022) Foliar application of dsRNA targeting endogenous potato (Solanum tuberosum) isoamylase genes ISA1, ISA2, ISA3 confers transgenic phenotype. Int J Mol Sci 24(1):190. https://doi.org/10.3390/ijms24010190
Singh A, Panwar R, Mittal P, Hassan MI, Singh IK (2021) Plant cytochrome P450s: Role in stress tolerance and potential applications for human welfare. Int J Biol Macromol 184:874–886. https://doi.org/10.1016/j.ijbiomac.2021.06.125
Singh A, Mehta S, Yadav S, Nagar G, Ghosh R, Roy A, Chakraborty A, Singh IK (2022) How to cope with the challenges of environmental stresses in the era of global climate change: An update on ROS stave off in plants. Int J Mol Sci 23(4):1995. https://doi.org/10.3390/ijms23041995
Sirohi P, Yadav BS, Afzal S, Mani A, Singh NK (2020) Identification of drought stress-responsive genes in rice (Oryza sativa) by meta-analysis of microarray data. J Genet 99(1):35. https://doi.org/10.1007/s12041-020-01195-w
Sun T, Zhang Y (2021) Short- and long-distance signaling in plant defense. Plant J 105(2):505–517. https://doi.org/10.1111/tpj.15068
Sun M, Shen Y, Chen Y, Wang Y, Cai X, Yang J, Jia B, Dong W, Chen X, Sun X (2022) Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol 189(4):2500–2516. https://doi.org/10.1093/plphys/kiac208
Swapnil P, Meena M, Rai AK (2021) Molecular interaction of nitrate transporter proteins with recombinant glycinebetaine results in efficient nitrate uptake in the cyanobacterium Anabaena PCC 7120. PLoS ONE 16(11):e0257870. https://doi.org/10.1371/journal.pone.0257870
Tang W, Thompson WA (2022) Role of the rice BAHD acyltransferase gene OsAt10 in plant cold stress tolerance. Plant Mol Biol Rep 40(3):482–499. https://doi.org/10.1007/s11105-021-01328-0
Taratima W, Chuanchumkan C, Maneerattanarungroj P, Trunjaruen A, Theerakulpisut P, Dongsansuk A (2022) Effect of heat stress on some physiological and anatomical characteristics of rice (Oryza sativa L) cv KDML105 callus and seedling. Biology 11(11):11. https://doi.org/10.3390/biology11111587
Thiranusornkij L, Thamnarathip P, Chandrachai A, Kuakpetoon D, Adisakwattana S (2018) Physicochemical properties of Hom Nil (Oryza sativa) rice flour as gluten free ingredient in bread. Foods 7(10):159. https://doi.org/10.3390/foods7100159
Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao N, Li X, Han C (2018) Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun 9(1):1063. https://doi.org/10.1038/s41467-018-03463-x
Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143(4):606–616. https://doi.org/10.1016/j.cell.2010.10.020
Umbreen S, Lubega J, Cui B, Pan Q, Jiang J, Loake GJ (2018) Specificity in nitric oxide signalling. J Exp Bot 69(14):3439–3448. https://doi.org/10.1093/jxb/ery184
Wang Y, Chen C, Loake GJ, Chu C (2010) Nitric oxide: Promoter or suppressor of programmed cell death? Protein Cell 1(2):133–142. https://doi.org/10.1007/s13238-010-0018-x
Wang WS, Pan YJ, Zhao XQ, Dwivedi D, Zhu LH, Ali J, Fu BY, Li ZK (2011) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L). J Exp Bot 62(6):1951–1960. https://doi.org/10.1093/jxb/erq391
Wang Y, Lin A, Loake GJ, Chu C (2013) H2O2 -induced leaf cell death and the crosstalk of reactive nitric/oxygen speciesF. J Integr Plant Biol 55(3):202–208. https://doi.org/10.1111/jipb.12032
Wang Y, Zhang Y, Fan C, Wei Y, Meng J, Li Z, Zhong C (2021) Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol 21(1):328. https://doi.org/10.1186/s12870-021-03083-6
Wang B, Yang X, Chen L, Jiang Y, Bu H, Jiang Y, Li P, Cao C (2022a) Physiological mechanism of drought-resistant rice co** with drought stress. J Plant Growth Regul 41(7):2638–2651. https://doi.org/10.1007/s00344-021-10456-6
Wang Y, Wang Y, Liu X, Zhou J, Deng H, Zhang G, **ao Y, Tang W (2022b) WGCNA analysis identifies the hub genes related to heat stress in seedling of rice (Oryza sativa L). Genes 13(6):1020. https://doi.org/10.3390/genes13061020
Withanawasam DM, Kommana M, Pulindala S, Eragam A, Moode VN, Kolimigundla A, Puram RV, Palagiri S, Balam R, Vemireddy LR, Varshney R (2022) Improvement of grain yield under moisture and heat stress conditions through marker-assisted pedigree breeding in rice (Oryza sativa L). Crop Pasture Sci 73(4):356–369. https://doi.org/10.1071/cp21410
Xu JH, Irshad F, Yan Y, Li C (2022) Loss of function of the RRMF domain in OsROS1a causes sterility in rice (Oryza sativa L). Int J Mol Sci 23(19):19. https://doi.org/10.3390/ijms231911349
Yang L, Li H (2023) Projecting the potential distribution and analyzing the bioclimatic factors of four Rhododendron subsect. Tsutsusi species under climate warming. J for Res 34:1707–1721. https://doi.org/10.1007/s11676-023-01626-1
Yang X, Wang B, Chen L, Li P, Cao C (2019) The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, quality. Sci Rep 9(1):3742. https://doi.org/10.1038/s41598-019-40161-0
Yang H, Sun Y, Wang H, Zhao T, Xu X, Jiang J, Li J (2021) Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against Stemphylium lycopersici in tomato. BMC Plant Biol 21(1):72. https://doi.org/10.1186/s12870-021-02848-3
Yang W, Chen Y, Gao R, Chen Y, Zhou Y, **e J, Zhang F (2023) MicroRNA2871b of Dongxiang wild rice (Oryza rufipogon Griff) negatively regulates cold and salt stress tolerance in transgenic rice plants. Int J Mol Sci 24(19):14502. https://doi.org/10.3390/ijms241914502
Yanhua C, Yaliang W, Huizhe C, **g X, Yikai Z, Zhigang W, Defeng Z, Yu** Z (2023) Brassinosteroids mediate endogenous phytohormone metabolism to alleviate high temperature injury at panicle initiation stage in rice. Rice Sci 30(1):70–86. https://doi.org/10.1016/j.rsci.2022.05.005
Yao D, Wu J, Luo Q, Zhang D, Zhuang W, **ao G, Deng Q, Bai B (2022) Effects of salinity stress at reproductive growth stage on rice (Oryza sativa L) composition, starch structure, physicochemical properties. Front Nutr 9:926217. https://doi.org/10.3389/fnut.2022.926217
Yuan X, Huang P, Wang R, Li H, Lv X, Duan M, Tang H, Zhang H, Huang J (2018) A zinc finger transcriptional repressor confers pleiotropic effects on rice growth and drought tolerance by down-regulating stress-responsive genes. Plant Cell Physiol 59(10):2129–2142. https://doi.org/10.1093/pcp/pcy133
Yurina NP (2023) Heat shock proteins in plant protection from oxidative stress. Mol Biol 57(6):951–964. https://doi.org/10.1134/S0026893323060201
Zehra A, Meena M, Dubey MK, Aamir M, Upadhyay RS (2017) Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot Stud 58:44. https://doi.org/10.1186/s40529-017-0198-2
Zehra A, Raytekar NA, Meena M, Swapnil P (2021) Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: A review. Curr Res Microb Sci 2:100054. https://doi.org/10.1016/j.crmicr.2021.100054
Zhang L, Huo Z, Amou M, **ao J, Cao Y, Gou P, Li S (2023a) Optimized rice adaptations in response to heat and cold stress under climate change in southern China. Reg Environ Change 23(1):25. https://doi.org/10.1007/s10113-022-02010-1
Zhang Q, Teng R, Yuan Z, Sheng S, **ao Y, Deng H, Tang W, Wang F (2023b) Integrative transcriptomic analysis deciphering the role of rice bHLH transcription factor Os04g0301500 in mediating responses to biotic and abiotic stresses. Front Plant Sci 14:1266242. https://doi.org/10.3389/fpls.2023.1266242
Zhou M, Tang W (2019) MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol Genet Genom 294(2):379–393. https://doi.org/10.1007/s00438-018-1516-4
Zhou X, Joshi S, Khare T, Patil S, Shang J, Kumar V (2021) Nitric oxide, crosstalk with stress regulators and plant abiotic stress tolerance. Plant Cell Rep 40(8):1395–1414. https://doi.org/10.1007/s00299-021-02705-5
Zhou H, Wang Y, Zhang Y, **ao Y, Liu X, Deng H, Lu X, Tang W, Zhang G (2022) Comparative analysis of heat-tolerant and heat-susceptible rice highlights the role of OsNCED1 gene in heat stress tolerance. Plants 11(8):1062. https://doi.org/10.3390/plants11081062
Zhu Y, Su H, Liu XX, Sun JF, **ang L, Liu YJ, Hu ZW, **ong XY, Yang XM, Bhutto SH, Li GB, Peng YY, Wang H, Shen X, Zhao ZX, Zhang JW, Huang YY, Fan J, Wang WM, Li Y (2024) Identification of NADPH oxidase genes crucial for rice multiple disease resistance and yield traits. Rice 17(1):1. https://doi.org/10.1186/s12284-023-00678-5
Zielonka J, Sikora A, Joseph J, Kalyanaraman B (2010) Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide. J Biol Chem 285(19):14210–14216. https://doi.org/10.1074/jbc.M110.110080
Zu X, Lu Y, Wang Q, La Y, Hong X, Tan F, Niu J, **a H, Wu Y, Zhou S, Li K, Chen H, Qiang S, Rui Q, Wang H, La H (2021) Increased Drought Resistance 1 mutation increases drought tolerance of upland rice by altering physiological and morphological traits and limiting ROS levels. Plant Cell Physiol 62(7):1168–1184. https://doi.org/10.1093/pcp/pcab053
Acknowledgements
The author P.S. gratefully acknowledges the financial support provided by the funding agency Science and Engineering Research Board (SERB), Empowerment and Equity Opportunities for Excellence in Science (File Number: EEQ/2022/000455). The author PS gratefully acknowledges the financial support provided by the funding agency DST-FIST (GP193). The author M.M. also gratefully acknowledges the financial support provided by the funding agency Science and Engineering Research Board (SERB), State University Research Excellence (SURE) (File Number: SUR/2022/005216) for this study. The author M.M. is highly thankful to the Ministry of Education and SPD-RUSA Rajasthan for the financial support received under the RUSA-2.0 project. All the authors acknowledge their host institute for infrastructure support. All the authors read and approved the content of the manuscript for the publication.
Funding
This research was funded by the funding agency Science and Engineering Research Board (SERB), State University Research Excellence (SURE) (File Number: SUR/2022/005216) and Science and Engineering Research Board (SERB), Empowerment and Equity Opportunities for Excellence in Science (File Number: EEQ/2022/000455). The research was also supported by the Ministry of Education and SPD-RUSA Rajasthan through the RUSA-2.0 project and the funding agency DST-FIST (GP193).
Author information
Authors and Affiliations
Contributions
A.S.P.S.: conceptualization, writing—original draft, writing draft—review and editing. P.P.: conceptualization, writing—original draft, writing draft—review and editing. R.B.: conceptualization, writing—original draft, writing draft—review and editing. A.Y.: conceptualization, writing—original draft, writing draft—review and editing. P.S.: conceptualization, research designed, investigation, methodology, writing—original draft, writing—review and editing, resources, project administration. C.S.S.: writing draft—review and editing. M. M.: investigation, conceptualization, research designed, methodology, resources, data curation, writing—original draft, writing draft—review and editing, project administration. All the authors reviewed and edited the contents. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors have their consent to publish their work.
Additional information
Communicated by Mansour Ghorbanpour.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abhijith Shankar, P.S., Parida, P., Bhardwaj, R. et al. Deciphering molecular regulation of reactive oxygen species (ROS) and reactive nitrogen species (RNS) signalling networks in Oryza genus amid environmental stress. Plant Cell Rep 43, 185 (2024). https://doi.org/10.1007/s00299-024-03264-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00299-024-03264-1