Abstract
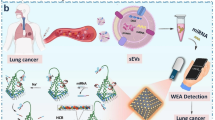
For the clinical diagnosis of diseases and for basic biological research, it is crucial to develop a trustworthy and efficient method for detecting small extracellular vesicles (sEVs) in multiple experimental conditions. Here, we create a colorimetric assay that enables sensitive and precise sEVs identification without the need for pricey equipment. In this assay, the exonuclease III (Exo III)-assisted signal recycle is activated by the released single-strand DNA (ssDNA) from SMBs (streptavidin magnetic beads)-aptamer-ssDNA complex after identification of sEVs. By integrating with the strand displacement amplification (SDA) process, a significant amount of double-strand DNA products with G-rich tails is produced. The G-rich tails fold to G-quadruplex under the assistance of hemin to catalyze the oxidation of TMB, yielding a color change. The approach offers a broad detection range of 5 orders of magnitudes based on the signal recycles and SDA. In addition, single-stranded DNA binding protein (SSB) is exploited in this method to minimize the background signal from non-specific digestion of Exo-III, making the method a robust tool for sEVs detection and disease diagnosis.
Similar content being viewed by others
Introduction
Cell culture supernatants and biofluids contain small extracellular vesicles (sEVs), a type of phospholipid-enclosed vesicle generated by parent cells (Malys et al. 2021; Moller and Lobb 2020; Urabe et al. 2020). sEVs are of relevance as disease indicators and therapeutic targets because they can transport proteins, nucleic acids, and other biological components to close or distant cells (Garcia-Martin et al. 2022; Zhao et al. 2022a; Niel et al. 2022). With pancreatitis and pancreatic cancer, which belong to the deadliest malignancy among pancreatic diseases, as example, the aberrant expression of sEVs is closely associated with cancer development (Armacki et al. 2020; Waldenmaier et al. 2021; Liu et al. 2021). Thus, the creation of a technique that can identify low abundance sEVs is important for the screening of cancers and helps to manage disease. In recent years, much efforts have been made to establish sensitive and reliable approaches for the sEVs detection. Among them, nanoparticle tracking analysis (NTA) has been employed extensively in clinical and experimental settings due to its superior capability in detecting trace amount of sEVs and characterizing the size distributions (Yahata et al. 2021; Zhao et al. 2020a, 2022b). However, the NTA method requires expensive equipment, complicated preparation, and laborious calculations, hampering the further application in non-laboratory sceneries (Zhao et al. 2020b, 2020c).
Colorimetric assays have attracted considerable attention recently because they allow for simple processing and direct readout of detection result with cheap and portable devices (**e et al. 2012; Marin et al. 2015; Yeasmin et al. 2020). In colorimetric assays, the presence of analytes in the sensing system lead to the color change that can be observed by naked eye and measured through optical instruments. For example, Yang et al. (2020) proposed a colorimetric bacteria detection approach based on the Exonuclease III (Exo-III) assisted stochastic DNA dual-walker for ultrafast target detection. Compared with the traditional approaches, the recently established colorimetric approaches possesses the advantages of low cost, fastness, and ease of working (Masumoto et al. 2022; Vogel et al. 2022). Therefore, the colorimetric assay can be potentially developed for sEVs detection. Despite that considerable progresses have been made among the former established colorimetric assays, the drawback of lacking enough sensitivity is inevitable in these colorimetric approaches. Various signal amplification strategies have been integrated in construction of the colorimetric assay to improve the sensitivity of DNA sequences detection, such as rolling circle amplification (RCA) (Hamidi and Perreault 2019), exponential amplification reaction (EXPAR) (Li et al. 2019), and strand displacement amplification (SDA) (Gong et al. 2021). Among them, SDA method have attracted abundant attention due to its high efficiency. However, the sensitivity and specificity of the SDA based approach need further improvement due to the limitation of threshold value in strand displacement. Exo-III is sequence-independent enzyme that can digests one strand of duplex DNA from a blunt end, 5′ overhang or nick (Zhao et al. 2022b; Wang et al. 2021). Exo-III has been widely exploited in constructing signal recycles to improve the sensitivity of current signal amplification approaches.
Inspired by the unique feature of Exo-III enzyme, we develop here a colorimetric approach for sensitive sEVs detection by integrating the Exo-III assisted signal recycles with SDA. In this method, a SMBs (streptavidin magnetic beads)-aptamer-ssDNA (single-strand DNA) complex is designed to specifically bind with target sEVs and mediate subsequent signal amplification. In the signal amplification process, a hairpin structure probe is designed and work as the activator of the amplification process through binding with released ssDNA sequences. The hybridization between released ssDNA sequences and activator probe initiates the Exo-III assisted signal recycles and SDA. The products after SDA process contains a G-rich sequence that can be fold to G-quadruplex under the assistance of hemin to catalyze the oxidation of TMB, yielding a color change. In addition, the single-stranded DNA binding protein (SSB), which can bind to single-stranded DNA and prevent ssDNA from being repaired to form double-stranded DNA or degraded by nucleases, is exploited in this approach to minimize the background signal from non-specific digestion of Exo-III, thus endowing the method a high specificity.
Experimental section
Materials and reagents
The oligonucleotides used in this colorimetric assay is listed in Additional file 1: Table S1. The sequences were synthesized and purified by Sangon Biotechnology Co. Ltd (Shanghai, China). Exonuclease III (Exo III) for sequence digestion and single-stranded DNA-binding protein (SSB) for reducing the background signal were obtained from New England Biolabs (Ipswich, MA, USA). Hydrogen peroxide (H2O2) and deoxynucleotide solution mixture (dNTPs) were obtained from TaKaRa Biotech. Inc. (Dalian, China). The color signals of the approach was monitored by the UV–visible spectrophotometer (UV-2550, Shimadzu, Kyoto, Japan).
Preparations of the probes
The synthesized activator probe was firstly diluted to 10 μM by 1 × PBS buffer solution. The mixture was heated to 90 °C for 10 min and was gradually cooled to room temperature. The assembled probes were stored at 4 °C for the subsequent experiments.
Detection procedures of the established approach
The Exo-III assisted signal recycles process was initiated by the addition of 2 μL sEVs (104 particles/μL), 2 μL MB probes (5 μM), 1 μL Exo-III enzyme (1 U/L), 2 μL H1 probes (5 μM), 2 μL H2 probes (5 μM). The mixture was diluted to 20 μL in volume by mixing with 11 μL buffer solution containing Tris–HCl (10 mM), NaCl (50 mM), MgCl2 (10 mM), and 100 μg/mL BSA. The 20 μL solution was incubated at 37 °C for 60 min. Afterward, 200 μL of ABTS2− (4 mM) and 1 μLH2O2 (30%) were added to the mixture, and the mixture was incubated at room temperature for 5 min. The absorbance of the mixture was monitored by UV–visible spectrophotometer (UV-2550, Shimadzu, Kyoto, Japan).
Data analysis
All data in this paper are expressed as mean ± standard deviations. Statistical significances between two groups were calculated by SPSS 22.0 by using two-tailed Student’s t test. The threshold for significance was set as P < 0.01.
Results and discussion
The working mechanism of the established approach for DNA sequences detection
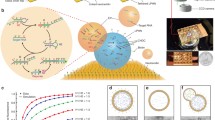
The working principle of the established colorimetric method is shown in Fig. 1. In this method, a SMBs-aptamer-ssDNA complex is designed, comprising CD63 aptamer, SMBs, and a ssDNA sequence that is partially complementary with CD63 aptamer. In the present of sEVs, the CD63 aptamer in the SMBs-aptamer-ssDNA complex precisely binds with CD63 protein on the surface of sEVs, leading to the liberation of ssDNA. After magnet based enrichment and isolation of the SMBs-aptamer-sEVs, the supernatant containing released ssDNA initiated the signal amplification process. In the signal amplification process, a hairpin structure probe is designed with a tail which is complementary with the ssDNA, and a DNAzyme section. In the present of released ssDNA, it binds with the tail in the activator probe, forming a blunt end in the probe. Under the assistance of the Exo-III enzyme, the stem section is digested from the 3’ terminal, resulting the liberation of the DNAzyme sequences and the ssDNA. The released ssDNA binds with a next activator probe to induce a signal recycle, and the DNAzyme sequence can bind with the loop section of the MB probe to form active secondary conformation. The active DNAzyme generates a nick in the loop section, leading to the release of the two single-strand DNA sequences (a and b). The b sequence, which is a homologous sequence with the ssDNA sequences, hybridizes with a next activator probe, inducing the second signal recycle; the a sequence binds with the H1 probe and gradually unfolds it to initiate the SDA process among the H1 probes and H2 probes. Eventually, the G-rich section in the H1 probe is exposed and can fold to active G quadruplex under the assistance of hemin to catalyze the oxidation of TMB, yielding a color change.
Construction of SMBs-aptamer-ssDNA complex and strategy to minimize the background signal of the established approach
To verify the construction of SMBs-aptamer-ssDNA complex and its capability in identifying sEVs, a fluorescence assay was performed through labeling FAM and corresponding quenching moiety (BHQ-1) on the terminal of CD63 aptamer and ssDNA, respectively. From the result in Fig. 2A, the signal of FAM labeled CD63 aptamer significantly decreased in the SMBs-aptamer-ssDNA complex, which was 5.67% of that in FAM labeled CD63 (2135.33 ± 78.5 a.u.), indicating that ssDNA was hybridized with CD63 aptamer and the SMBs-aptamer-ssDNA complex was successfully constructed. Upon the addition of sEVs, the FAM signal recovered, implying the ssDNA disassociated from SMBs-aptamer-ssDNA complex.
One of the biggest challenge for the Exo-III based approaches is high background signal derived from the ono-specific digestion of probes by Exo-III. Herein, we exploited the single-stranded DNA binding protein (SSB) which possesses a high affinity to the single-strand DNA sequences (ssDNA), to reduce the background signals. To demonstrate the efficiency of the SSB to minimize the background signal, a fluorescence assay was performed. In this fluorescence assay, the two terminals of the activator probe were labeled with FAM and BHQ-1, respectively. If the activator probe was digested by the Exo-III, fluorescence signal of FAM would reappeared. As shown in Fig. 2B, the fluorescence intensity in the solution containing only activator probe and the synthesized ssDNA sequence (control group) was 5.65 a.u.. The intensities of the group containing Exo-III enzyme and activator probe gradually elevated with the time ranged from 0 to 60 min, and the intensity at 60 min was approximate 2.1 times higher than that in the control group. On the contrary, the intensity of the group containing Exo-III enzyme, SSB and activator probe showed no obvious increment compared with the control group.
Feasibility of the colorimetric approach
To test the feasibility of the established colorimetric assay for sensitive sEVs detection, the color change and quantitative absorbance of the approach under different experimental conditions was monitored. From the result in Fig. 3, no any color change was observed when the sEVs was absent, indicating the Exo-III alone cannot induce signal recycles and color generation. Similar results were obtained when MB, H1 probe and H2 probe absent in the sensing system, implying all the probes were essential for the feasibility of the colorimetric approach. When all essential experimental components existed in this system, an obvious color change was observed.
Optimization of experimental conditions
Several experimental conditions can affect the detection performance of the established colorimetric approach, including the concentration of Exo-III enzyme, the concentration of MB. As a decisive parameter in constructing the signal recycles, the amount of Exo-III in the sensing system determines the efficiency of digestion. Therefore, we firstly investigated the absorbance of the approach when different concentrations of Exo-III were exploited in the system. From the result in Fig. 4A, the absorbance of the system enhanced with the concentration of Exo-III ranged from 0 to 1 U/L, and no more increasements were observed when more than 1U/L Exo-III was added in the system. Therefore, 1U/L Exo-III was selected for the subsequent experiments. Meanwhile, the optimized concentration of MB was 200 nM (Fig. 4B).
Analytical performance of the established colorimetric approach
To evaluate the detection performance of the established approach, we have recorded the color of each group and monitored the absorbance of the method when detecting different concentrations of sEVs. From the result in Fig. 5A, a gradually darkening color was observed when the concentration of sEVs increased. The obtained absorbance of the approach increased with the concentration of DNA sequences ranged from 103 particles/μL to 107 particles/μL. The correlation equation between the absorbance and the concentration of DNA sequences was determined Y = 0.1250*lgC − 0.003, with the correlation coefficient of 0.9786 (Fig. 5B). Meanwhile, the approach possesses a wide detection rage of 5 orders of magnitude and low limit of detection (LOD) of 267 particle/μL (LOD was calculated based on the 3σ/slope method). Compared with the former sEVs detection methods, the established approach showed a comparable detection sensitivity and a wider detection range (Additional file 1: Table S2). To test the specificity of the established approach, the method was applied for the detection of interferents, including ATP (adenosine triphosphate), CEA (carcino-embryonic antigen), CRP (C-reactionprotein). From the result in Fig. 5C, the absorbance of the approach when detecting interferents showed no significant differences with the control group. An obvious color change was observed when detecting sEVs. These above experimental results proved the high selectivity of the proposed approach in discrimination of sEVs from the Interference analytes. We then performed a recovery assay to study whether the proposed approach could be utilized in detecting sEVs in complicated biological samples. In the recovery assay, different concentrations of sEVs were added in the 5% and 10% artificial serum samples, and the recovery rate was calculated as shown in Table 1. Consequently, a quantitative recovery rate ranged from 99.1 to 106.6% were obtained, which is comparable or superior to most of the existing Exo-III based approaches.
Conclusion
In summary, we depict here a novel colorimetric method for sensitive and accurate sEVs detection by integrating Exo-III assisted signal recycles and SDA mediated color generation. Based on the elegant design, the method exhibited a wide detection range of 5 orders of magnitudes, which is superior or comparable to most of the former reported colorimetric sEVs detection assays. In addition, a minimized background signal was obtained by exploiting the SSB to reduce the ono-specific digestion of ssDNA by Exo-III enzyme. Moreover, this method shows a robust capability in discriminating sEVs from interferents in complicated biological samples. All the results indicate the great potential of the approach in clinical application and disease diagnosis.
Availability of data and materials
Almost all details of experimental data are presented in the article or additional file.
References
Armacki M, Polaschek S, Waldenmaier M, Morawe M, Ruhland C, Schmid R, Lechel A, Tharehalli U, Steup C, Bektas Y, Li H, Kraus JM, Kestler HA, Kruger S, Ormanns S, Walther P, Eiseler T, Seufferlein T. Protein kinase D1, reduced in human pancreatic tumors, increases secretion of small extracellular vesicles from cancer cells that promote metastasis to lung in mice. Gastroenterology. 2020;159(3):1019-1035.e22.
Garcia-Martin R, Wang G, Brandao BB, Zanotto TM, Shah S, Kumar Patel S, Schilling B, Kahn CR. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601(7893):446–51.
Gong S, Zhang S, Wang X, Li J, Pan W, Li N, Tang B. Strand displacement amplification assisted CRISPR-Cas12a strategy for colorimetric analysis of viral nucleic acid. Anal Chem. 2021;93(45):15216–23.
Hamidi SV, Perreault J. Simple rolling circle amplification colorimetric assay based on pH for target DNA detection. Talanta. 2019;201:419–25.
Li R, Liu Q, ** Y, Li B. G-triplex/hemin DNAzyme: an ideal signal generator for isothermal exponential amplification reaction-based biosensing platform. Anal Chim Acta. 2019;1079:139–45.
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, Fu W, Yi J, Wang J, Du G. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11(9):2783–97.
Malys MSS, Aigner C, Schulz SMM, Schachner H, Rees AJJ, Kain R. Isolation of small extracellular vesicles from human sera. Int J Mol Sci. 2021;22(9):4653.
Marin MJ, Schofield CL, Field RA, Russell DA. Glyconanoparticles for colorimetric bioassays. Analyst. 2015;140(1):59–70.
Masumoto M, Ohta S, Nakagawa M, Hiruta Y, Citterio D. Colorimetric paper-based sarcosine assay with improved sensitivity. Anal Bioanal Chem. 2022;414(1):691–701.
Moller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer. 2020;20(12):697–709.
Urabe F, Kosaka N, Ito K, Kimura T, Egawa S, Ochiya T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am J Physiol Cell Physiol. 2020;318(1):C29–39.
van Niel G, Carter DRF, Clayton A, Lambert DW, Raposo G, Vader P. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat Rev Mol Cell Biol. 2022;23(5):369–82.
Vogel U, Beerens K, Desmet T. A colorimetric assay for the screening and kinetic analysis of nucleotide sugar 4,6-dehydratases. Anal Biochem. 2022;655:114870.
Waldenmaier M, Seibold T, Seufferlein T, Eiseler T. Pancreatic cancer small extracellular vesicles (exosomes): a tale of short- and long-distance communication. Cancers (basel). 2021;13(19):4844.
Wang N, Jiang YJ, Zhang X, Lin HR, Cheng F, Li Q, Li CM, Huang CZ. Nanosurface energy transfer indicating Exo III-propelled stochastic 3D DNA walkers for HIV DNA detection. Analyst. 2021;146(5):1675–81.
**e X, Xu W, Liu X. Improving colorimetric assays through protein enzyme-assisted gold nanoparticle amplification. Acc Chem Res. 2012;45(9):1511–20.
Yahata S, Hirose M, Ueno T, Nagumo H, Sakai-Kato K. Effect of sample concentration on nanoparticle tracking analysis of small extracellular vesicles and liposomes mimicking the physicochemical properties of exosomes. Chem Pharm Bull (tokyo). 2021;69(11):1045–53.
Yang H, **ao M, Lai W, Wan Y, Li L, Pei H. Stochastic DNA dual-walkers for ultrafast colorimetric bacteria detection. Anal Chem. 2020;92(7):4990–5.
Yeasmin S, Ammanath G, Ali Y, Boehm BO, Yildiz UH, Palaniappan A, Liedberg B. Colorimetric urinalysis for on-site detection of metabolic biomarkers. ACS Appl Mater Interfaces. 2020;12(28):31270–81.
Zhao X, Luo C, Mei Q, Zhang H, Zhang W, Su D, Fu W, Luo Y. Aptamer-cholesterol-mediated proximity ligation assay for accurate identification of exosomes. Anal Chem. 2020a;92(7):5411–8.
Zhao X, Zeng L, Mei Q, Luo Y. Allosteric probe-initiated wash-free method for sensitive extracellular vesicle detection through dual cycle-assisted CRISPR-Cas12a. ACS Sens. 2020b;5(7):2239–46.
Zhao X, Zhang W, Qiu X, Mei Q, Luo Y, Fu W. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal Bioanal Chem. 2020c;412(3):601–9.
Zhao H, Chen X, Hu G, Li C, Guo L, Zhang L, Sun F, **a Y, Yan W, Cui Z, Guo Y, Guo X, Huang C, Fan M, Wang S, Zhang F, Tao L. Small extracellular vesicles from brown adipose tissue mediate exercise cardioprotection. Circ Res. 2022a;130(10):1490–506.
Zhao X, Yuan Y, Liu X, Mao F, Xu G, Liu Q. A versatile platform for sensitive and label-free identification of biomarkers through an exo-III-assisted cascade signal amplification strategy. Anal Chem. 2022b;94(4):2298–304.
Acknowledgements
The authors thank financial and technical support from the Natural Science Foundation of Chongqing Science and Technology Commission (cstc2021jcyj-msxmX0916) and the Natural Science Foundation of Chongqing Medical and Pharmaceutical College (ygz2019302 and ygz2019305).
Funding
No fund available.
Author information
Authors and Affiliations
Contributions
FY and HC is the supervisor of the team in all research steps including designing, data analysis, and manuscript writing. LN, as the first author, has the main role for experimental data collection, data gathering, preparation of results, and data analysis. YZ, YX, and ZD assist the data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The manuscript does not contain clinical or trial studies on patients, humans, or animals.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Table S1.
Details of the sequences used in this research, Supporting information. Table S2. Comparison of the established approach with former ones.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ning, L., Zhou, Y., **e, Y. et al. Ultrasensitive and colorimetric small extracellular vesicles (sEVs) analysis via dual-cycle signal tool. J Anal Sci Technol 14, 5 (2023). https://doi.org/10.1186/s40543-023-00370-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40543-023-00370-9