Abstract
Background
Even though the roles of pentatricopeptide repeat (PPR) proteins are essential in plant organelles, the function of many chloroplast-targeted PPR proteins remains unknown. Here, we characterized the function of a chloroplast-localized PPR protein (At3g59040), which is classified as the 287th PPR protein among the 450 PPR proteins in Arabidopsis (http://ppr.plantenergy.uwa.edu.au).
Results
The homozygous ppr287 mutant with the T-DNA inserted into the last exon displayed pale-green and yellowish phenotypes. The microRNA-mediated knockdown mutants were generated to further confirm the developmental defect phenotypes of ppr287 mutants. All mutants had yellowish leaves, shorter roots and height, and less seed yield, indicating that PPR287 is crucial for normal Arabidopsis growth and development. The photosynthetic activity and chlorophyll content of ppr287 mutants were markedly reduced, and the chloroplast structures of the mutants were abnormal. The levels of chloroplast rRNAs were decreased in ppr287 mutants.
Conclusions
These results suggest that PPR287 plays an essential role in chloroplast biogenesis and function, which is crucial for the normal growth and development of Arabidopsis.
Similar content being viewed by others
Background
The chloroplast genome encodes approximately 120–130 proteins necessary for photosynthesis and plastid biogenesis [1,2,3]. However, more than 3000 nucleus-encoded proteins are transported into the chloroplast, many of which are needed for chloroplast gene expression [4,5,6,7]. Chloroplast gene expression is modulated and regulated by posttranscriptional processes, such as mRNA and tRNA splicing, mRNA editing, RNA stability, and translational control, during which many RNA-binding proteins (RBPs) play essential roles [8,9,10,11,12]. In particular, most of the RBPs involved in chloroplast RNA metabolism are nucleus-encoded and are transported into chloroplasts [5,6,7, 13, 14].
The pentatricopeptide repeat (PPR) proteins are among the nucleus-encoded chloroplast RBPs. Notably, PPR proteins are particularly abundant in land plants [15,16,17]. The Arabidopsis genome encodes more than 450 PPR proteins, whereas less than 10 PPR proteins are found in humans [15, 18], suggesting their plant-specific functions. PPR proteins are divided into several subfamilies in terms of their tandem repeat motifs and additional domains. In general, the P-class PPR proteins that harbor only the PPR motifs are involved in intercistronic processing, splicing of group II introns, and RNA stabilization, whereas the PLS-class PPR proteins that contain additional C-terminal domains, such as E, E+, and DYW, are required for C to U RNA editing [17, 19, 20].
In chloroplasts of several plant species, including Arabidopsis thaliana, rice (Oryza sativa), and maize (Zea mays), the functions of many PPR proteins have been determined, and most of the chloroplast PPR proteins characterized so far participate mainly in the stabilization of mRNAs. Examples include CRP1 for stabilizing the 5′ and 3′ ends of the petB-petD intergenic region in maize [21, 22], PPR10 for stabilizing the 5′ and 3′ ends of the atpI-atpH and psaJ-rpl33 intergenic regions in maize [23,24,25], HCF152 for stabilizing the 5′ and 3′ ends of the psbH-petB intergenic region in Arabidopsis thaliana [26, 27], MRL1 for stabilizing the rbcL 5′ end in Arabidopsis [28], and PGR3 for stabilizing the petL 5′ end in Arabidopsis [29, 30]. Similarly, mitochondrial PPR19 and MTSF1 are needed for stabilizing the nad1 intron 3′ end and nad4 3′ end in Arabidopsis [31, 32]. Analysis of loss-of-function mutants demonstrated that these aforementioned PPR proteins are critical for normal growth and development of plants. Although these previous studies clearly show that PPR proteins are essential for organellar functions and plant development, the cellular functions of many PPR proteins still remain to be characterized.
In this study, we determined the function of a chloroplast-targeted PPR protein possessing 10 PPR motifs (At3g59040), which is classified as the 287th PPR protein among the 450 PPR proteins in Arabidopsis (http://ppr.plantenergy.uwa.edu.au) [33], thus designated PPR287. We show that PPR287 affects the level of chloroplast rRNAs, which is essential for chloroplast biogenesis and function as well as for the normal growth and development of Arabidopsis.
Results
Cellular localization and expression patterns of PPR287
PPR287 harbors a putative chloroplast transit peptide at the N-terminus, ten tandemly repeated PPR motifs comprising from 146 to 495 amino acids, and a region rich in aspartate, glutamate, leucine, and serine residues at the C-terminus (Fig. 1a). To verify whether PPR287 is indeed transported into chloroplasts, transgenic Arabidopsis plants that express the PPR287-green fluorescent protein (GFP) fusion protein or only the GFP protein as a control were generated and analyzed by means of confocal microscopy. Evidently, GFP signals were observed in the chloroplasts of PPR287-GFP expressing transgenic plants (Fig. 1b), whereas GFP signals were detected in the nucleus and cytoplasm of GFP-expressing transgenic plants (Additional file 1), indicating that the nucleus-encoded PPR287 is transported into chloroplasts. To identify the tissue-specific expression patterns of PPR287, an approximately 1.5-kb fragment of the genomic DNA harboring the putative promoter of PPR287 was cloned in front of a GUS reporter gene, and the PPR287PRO::GUS expression was observed in transgenic Arabidopsis plants. Strong GUS signals were detected in young seedlings, leaves, and flowers, whereas only weak GUS signals were observed in stems and siliques (Additional file 2).
The domain structure and cellular localization of PPR287. a Schematic representation of the PPR287 domain structures. The chloroplast transit peptide (cTP) and PPR motifs are shown. The number of amino acid (aa) in PPR287 is indicated, and a region rich in aspartate (D), glutamate (E), leucine (L), and serine (S) residues at the C-terminus is indicated. b Cellular localization of the PPR287 protein. GFP signals from the PPR287-GFP-expressing transgenic Arabidopsis were observed using a confocal microscope. Red signals indicate chloroplast auto-fluorescence. Bar = 10 μm
PPR287 is essential for normal growth and development of Arabidopsis
To identify the function of PPR287 in plant growth and development, two T-DNA insertion Arabidopsis mutant lines were obtained and analyzed. The CS814021 line has T-DNA inserted into the second exon (Fig. 2a); about a fourth of its seeds were aborted and shriveled (Fig. 2d). Importantly, the homozygous mutant of the CS814021 line could not be obtained.
Development-defect phenotypes of the ppr287 mutants. a Schematic representation of T-DNA insertion sites and amiRNA knockdown mutant (KD1) target site. Black rectangles and lines represent exons and introns, respectively, and untranslated regions are indicated by white rectangles. The positions of T-DNA insertion in the ppr287 mutants and amiR target site are indicated by black and white triangles, respectively. The number of amino acid (aa) in PPR287 is indicated. Positions of primers used for RT-PCR in (b and c) are shown with arrowheads. b RT-PCR analysis confirming the absence or downregulation of PPR287 expression in the ppr287 mutant, amiR mutant (KD1), and complementation line (Com1) in ppr287 mutant background. The primer pair (1 + 2) amplifying the PPR287 transcript spanning the T-DNA insertion site was used. c RT-PCR (left) and quantitative real-time RT-PCR (right) analyses confirming the downregulation of PPR287 expression in the ppr287 mutant. The primer pair (3 + 4) amplifying the PPR287 transcript upstream of the T-DNA insertion site was used. Tubulin was used as a loading control. d Seed development of the heterozygous mutant. White arrows indicate aborted seeds in the mutants. Bar = 1 cm. e Pale-green phenotypes of the ppr287 and KD1 line observed at 6 days after germination
All CS814021 mutant seeds that were germinated on MS medium containing sucrose were heterozygotes. Many CS814021 mutant seeds were not germinated on sucrose-containing MS medium, which are supposed to be homozygotes. These results suggest that PPR287 is essential for embryogenesis. In contrast, a homozygous mutant of the SALK_041236C line, in which T-DNA was inserted at the position encoding 572th amino acid near the stop codon in the last exon of PPR287 (Fig. 2a), could be obtained. No band corresponding to PPR287 was detected in the ppr287 mutant by RT-PCR using the primer pair amplifying the PPR287 transcript spanning the T-DNA insertion site (Fig. 2b), suggesting knockout mutant. However, because the T-DNA is inserted downstream of the PPR motif (Fig. 2a), it is possible that partially truncated PPR287 protein containing entire PPR motifs can be expressed in the ppr287 mutant, suggesting the ppr287 mutant is not a genuine loss-of-function mutant. To examine whether T-DNA insertion affects the transcript level of PPR287 in the mutant, the level of PPR287 in the mutant was analyzed by RT-PCR and quantitative real-time RT-PCR using the primer pair amplifying the PPR287 transcript upstream of the T-DNA insertion site (Fig. 2a). Cleary, the PPR287 level was decreased down to approximately 50% of the wild-type level (Fig. 2c), confirming that the ppr287 is a knockdown mutant.
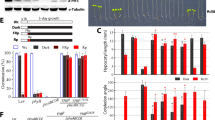
Because the loss-of-function of PPR287 leads embryo lethality, we generated ppr287 knockdown mutants by means of an artificial microRNA (amiRNA)-mediated knockdown method. The transgenic Arabidopsis plants expressing the amiRNA, which was designed to cleave the first exon of PPR287, were generated, and downregulation of PPR287 in each transgenic line was confirmed by RT-PCR and real-time RT-PCR analysis (Fig. 2b and Additional file 3). The amiRNA knockdown (KD) lines as well as the ppr287 mutant displayed development-defect phenotypes, such as yellowish and pale-green leaves (Fig. 2e). Interestingly, the severity of developmental defects was closely correlated to the knockdown levels of PPR287; the KD1 line, in which the PPR287 level was decreased down to approximately 30% of the wild-type level, exhibited the most severe abnormal development, whereas the KD2 and KD3 lines, in which PPR287 levels were approximately 40 to 60% of the wild-type levels, showed mild abnormal development (Additional file 3). Among the three knockdown lines, the KD1 line showing obvious phenotypes was chosen for further analysis. To further confirm the function of PPR287, a complementation line expressing PPR287 in the ppr287 mutant background (Com1/ppr287) was also generated and analyzed. The expression of PPR287 in the complementation line (Com1) was confirmed by RT-PCR (Fig. 2b). Abnormal development was observed in both the knockout ppr287 and the knockdown KD1 mutants, whereas the complementation line exhibited wild-type phenotypes (Fig. 2e). Germination of the mutant seeds was slightly retarded (Fig. 3a), and root growth of the mutants was also more inhibited than that of the wild type (Fig. 3b). The delayed growth and development of the mutants were much more evident when grown in soil (Fig. 3c and d). The plantlets and leaves of the mutants were much smaller than those of the wild type (Additional file 4). Bolting time was delayed in the mutants, but the number of leaves at bolting was not different between the wild-type and mutant plants (Additional file 4), suggesting that PPR287 does not affect flowering time. Plant height and seed yield were significantly decreased in the mutants compared with the wild type (Additional file 4). Collectively, these results suggest that PPR287 is essential for normal growth and development of Arabidopsis.
Growth-defective phenotypes of the ppr287 mutant. a Seed germination rates of the Col-0, ppr287 mutant, amiRNA knockdown mutant (KD1), and complementation line (Com1) were analyzed on the indicated days. b Root lengths of the plants were measured 10 days after germination (DAG). Data are mean ± SD of three biological replicates (n = 5). Bar = 1 cm. c Growth of the plants at 20 DAG. Bar = 1 cm. d Height of the plants at 50 DAG. Bar = 10 cm
PPR287 affects photosynthetic activity and chloroplast biogenesis
Because ppr287 mutants showed retarded growth and pale-green phenotypes (Fig. 4a), we investigated whether PPR287 affects photosynthetic activity, chlorophyll content, and chloroplast biogenesis. It was evident that photosynthetic activity, as shown by maximum quantum yield of photosystem II (Fv/Fm), was significantly reduced in ppr287 mutants compared with the wild type (Fig. 4b). Chlorophyll contents in the mutants were much less than those in the wild type (Fig. 4c). Photosynthetic activity and chlorophyll content of the complementation line recovered to wild-type levels. These results suggest that PPR287 plays an essential role in photosynthetic activity and chlorophyll biosynthesis.
PPR287 is crucial for photosynthesis and chloroplast biogenesis. a Pale-green phenotypes of the ppr287 mutant and amiRNA knockdown mutant (KD1) observed at 20 days after germination (DAG). Bar = 1 cm. b Chlorophyll fluorescence (Fv/Fm) and (c) chlorophyll contents in each plant at 10 DAG. Data are mean ± SD of three biological replicates (n = 5), and statistically different values are indicated by asterisks (p ≤ 0.05). d Chloroplast structures in the leaves of 3-week-old plants were observed using a transmission electron microscope. Bar = 1 μm. e In situ detection of H2O2 levels in the leaves of each plant were performed by DAB (3,3′-diaminobenzidine) staining. Bar = 5 mm. The levels of H2O2 were quantified, and data are mean ± SD of three biological replicates (n = 5). FW, fresh weight
We next examined whether PPR287 is involved in chloroplast biogenesis by observing chloroplast structures using a transmission electron microscope. Chloroplast morphology was abnormal in ppr287 mutants, displaying a few and loosely stacked thylakoids, whereas the wild type and the complementation line had normal chloroplasts (Fig. 4d and Additional file 5). To identify whether this abnormal chloroplast structure affects chloroplast function, we analyzed the levels of reactive oxygen species (ROS), which are an indicator of chloroplast function. Among ROS, such as superoxide (O2−), hydrogen peroxide (H2O2), and nitric oxide (NO), the amount of H2O2 was measured using a 3,3′-diaminobenzidine (DAB) staining. Evidently, ppr287 mutants had much stronger DAB staining than the wild type and complementation line (Fig. 4e). These results were further supported by quantifying H2O2 levels in the leaves of each plant. The levels of H2O2 were much higher in ppr287 mutants than the wild type and complementation line (Fig. 4e). Collectively, these results imply that PPR287 is important for photosynthesis and chloroplast biogenesis and function.
PPR287 affects transcript levels of chloroplast rRNAs
The next important task is to determine how PPR287 affects chloroplast biogenesis and function. Because diverse PPR proteins are involved in organellar RNA metabolism, including intron splicing and RNA stability, we first analyzed whether PPR287 is involved in the splicing of chloroplast introns. The results showed that the splicing efficiencies of none of the intron-containing chloroplast mRNAs and tRNAs were altered in ppr287 mutants (Additional file 6), suggesting that PPR287 is not involved in the splicing of chloroplast introns. Next, we analyzed whether PPR287 affects the expression of chloroplast genes. The levels of none of the chloroplast transcripts were altered in ppr287 mutants (Additional file 7), suggesting that PPR287 does not affect the expression of chloroplast genes. Finally, we investigated whether PPR287 is involved in chloroplast rRNA stability by northern blot analysis. The precursor and mature products of 4.5S, 5S, 16S, and 23S rRNAs were detected with the probes specific to each rRNA. The results showed that the levels of all rRNAs were decreased in ppr287 mutants. Notably, both the precursor and mature products of 23S rRNA were decreased in the mutants (Fig. 5). The decreased intensities of rRNA products observed in ppr287 mutants recovered to normal level in the complementation line. These results suggest that PPR287 may affect rRNAs stability in chloroplasts.
Decreased levels of chloroplast rRNAs in the ppr287 mutant. Total RNA extracted from 1-week-old Col-0, ppr287 mutant, amiRNA knockdown mutant (KD1), and complementation line (Com1) was separated on a 1.2% formaldehyde agarose gel, and the levels of the processed products were determined by Northern blot analysis using the probes represented by thick lines below each gene; three probes for rrn23 are labeled with (a), (b), and (c). Relative intensities of rRNA bands in each sample were calculated, and the values under each lane are mean ± SD of three replicates
Discussion
Contrary to the increasing understanding of the distribution and organellar targeting of PPR proteins in higher plants, the function and cellular role of only a few PPR proteins have been determined until recently. Our current results demonstrate that PPR287 plays a crucial role in Arabidopsis development by affecting the level of chloroplast rRNAs. No loss-of-function homozygous mutants of Arabidopsis PPR287 could be obtained (Fig. 2), suggesting that PPR287 is essential for plant development. In contrast, the homozygous mutant of PPR287 that has T-DNA inserted near the stop codon in the last exon of PPR287 (Fig. 2) could be obtained, which showed pale-green phenotypes but developed normal seeds (Fig. 2). This mutant presumably behaves like a knockdown mutant because it can synthesize partially truncated functional proteins (Fig. 2). This hypothesis was further supported by the observation that ppr287 knockdown mutants generated by an amiRNA knockdown method exhibited similar pale-green phenotypes. Both knockdown mutants displayed severe defects in leaf greening, photosynthesis, chlorophyll biosynthesis, and chloroplast biogenesis (Figs. 2, 3 and 4).
Notably, PPR287 affects transcript levels of chloroplast rRNAs, which then affects chloroplast biogenesis and function. When the processing of rRNAs is impaired, the mature rRNAs are decreased, with concomitant increase in the precursor rRNAs, as exemplified in other PPR mutants, such as svr7 and sot1 [34, 35]. Because our northern blotting analysis revealed that the intensities of precursor rRNAs were not increased in ppr287 mutants, but both the precursor and mature products of all chloroplast rRNAs were decreased in the mutants (Fig. 5a), we propose that PPR287 affects the stability of chloroplast rRNAs. Although we cannot rule out the possibility that the lower amount of rRNAs in ppr287 mutants is due to the lower transcription of rRNA genes, it is unlikely that PPR287, as an RNA-binding protein, affects transcription of rRNA genes. Contrary to many reports demonstrating the roles of PPR proteins in the stabilization of chloroplast mRNAs, the function of PPR proteins involved in the stabilization of rRNAs is largely unknown. A previous study has shown that maize PPR53, which is orthologous to the Arabidopsis PPR protein SOT1, enhances the stability of chloroplast 23S rRNA [36]. Our current results add PPR287 as another PPR protein possibly involved in the stabilization of chloroplast rRNAs. However, we do not know at present the mechanistic role of PPR287 affecting the level of chloroplast rRNAs. Given that plastid rRNA operon encodes all plastid rRNAs and two tRNAs (trnI and trnA) as a single transcript unit (Fig. 5), which is then processed to each mature transcript, it is likely that PPR287 binds to the primary or precursor rRNA transcript and thereby stabilizes all chloroplast rRNAs (Fig. 5). More analysis is required to identify the binding sites of PPR287 and the effects of PPR287 binding on the stabilization of chloroplast rRNAs. In particular, it would be necessary to determine whether PPR287 binds to the specific sequence in the 5′ end or 3′ end of plastid primary rRNA transcript and thereby affects the stability of all rRNAs or whether PPR287 stabilizes all chloroplast rRNAs by binding to the conserved sequences in each rRNA transcript. In addition, it would be interesting to investigate whether PPR287 interacts with other proteins, which affects the level and stability of chloroplast rRNAs. These key experiments will greatly contribute to fully understand the cellular role of PPR287 in the stabilization of chloroplast rRNAs.
Conclusions
Our results demonstrate that the chloroplast-transported PPR287 affects transcript levels of chloroplast rRNAs, which is crucial for chloroplast biogenesis and function during plant growth and development. Given that the role of PPR proteins in the stabilization and processing of chloroplast rRNAs has been identified in only a few cases, our results demonstrating that PPR287 affects the level of all chloroplast rRNAs are intriguing. Characterizing the effect of PPR287 binding on the stabilization of chloroplast rRNAs should be a next important experiment. Moreover, further research is needed to identify the functions of many as-yet uncharacterized PPR proteins and their coordinated roles in the splicing, processing, and stability of chloroplast transcripts.
Methods
Plant materials, mutants, and growth conditions
The wild-type and mutant A. thaliana were Col-0 ecotype. The Arabidopsis T-DNA insertion mutants, CS814021 and SALK_ 041236C, were obtained from the Arabidopsis Biological Resources Center. The ppr287 knockdown mutants were generated using an artificial microRNA-mediated knockdown method [37, 38]. The amiRNA constructs targeting the first exon of PPR287 were designed using the Web MicroRNA Designer program (http://wmd3.weigelworld.org) and were cloned into the pBI121 vector. Arabidopsis transformation was performed by means of vacuum infiltration using Agrobacteruim tumefaciens GV3101 [39]. Complementation lines were generated by expressing the full-length PPR287 under the control of the cauliflower mosaic virus 35S promoter in the ppr287 mutant background. The T3 or T4 homozygote transgenic lines were selected for phenotypic analysis. The expression levels of PPR287 in knockdown mutants and complementation lines were analyzed by RT-PCR with gene-specific primers listed in Additional file 8. All plants were grown in soil or half-strength Murashige and Skoog (MS) medium containing 1% sucrose at 23 ± 2 °C under long-day conditions (16 h-light / 8 h-dark cycle).
Analysis of subcellular localization of PPR287
The cDNA encoding full-length PPR287 was cloned into the XbaI/EcoRI site of CsV-GFP3-PA vector using the primers listed in Additional file 8, which expresses the PPR287-GFP fusion protein under the control of cassava vein mosaic virus promoter. Transgenic Arabidopsis plants that express the PPR287-GFP fusion protein were generated by means of vacuum infiltration using Agrobacteruim tumefaciens GV3101 [39], and GFP signals were detected using a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss Inc. Thornwood, NY, USA). The excitation and emission wavelengths were 488 and 505 nm, respectively.
Chlorophyll content and chlorophyll fluorescence measurement
Chlorophyll content was measured using ethanol extraction and quantification method as previously described [40]. Briefly, leaves of one-week-old wild-type, mutants, and complementation lines were ground in liquid nitrogen, and chlorophyll was extracted with 96% ethanol. The samples were kept overnight at room temperature in the dark. After centrifugation, the absorbance of the supernatant was measured at 648 nm and 664 nm. Chlorophyll fluorescence (Fv/Fm) was measured with a Handy PEA chlorophyll fluorimeter according to the manufacturer’s instructions (Hansatech Instruments Ltd., Norfolk, UK).
Transmission electron microscopy
Chloroplast structures were analyzed using TEM as previously described [41, 42]. Briefly, two-week-old seedlings were fixed with a mixture of 2% glutaraldehyde and 2% paraformaldehyde in 50 mM cacodylate buffer, pH 7.2, at room temperature for 4 h. The samples were embedded in LR White (London Resin Co., London, UK) at 50 °C for 24 h, and thin sections (80–100 nm thickness) were prepared using an ultra-microtome with a diamond knife. The thin sections were stained with uranyl acetate and lead citrate and then examined using a transmission electron microscope JEM-1400 (Jeol, Tokyo, Japan).
RNA extraction, RT-PCR, and northern blot analysis
Total RNA was isolated from the frozen samples using the Plant RNeasy extraction kit (Qiagen, Valencia, CA, USA). The splicing pattern of the intron-containing genes was analyzed by RT-PCR using the gene-specific primers listed in Additional file 8 as previously described [43]. Splicing efficiency was measured by real-time RT-PCR using the gene-specific primers listed in Additional file 9 as previously described [32, 41]. The levels of chloroplast transcripts were measured by quantitative RT-PCR using the gene-specific primers listed in Additional file 10. Real-time RT-PCR was carried out on a Rotor-Gene Q thermal cycler (Qiagen) using a SYBR Green RT-PCR kit (Qiagen). For northern blot analysis, four or five micrograms of total RNA were separated on a 1.2% formaldehyde-agarose gel and transferred to a Hybond-N+ nylon membrane (Amersham Biosciences, Parsippany, NJ, USA). The [α-32P]-labeled probes were synthesized using a random primer DNA labeling kit (TaKaRa Bio., Shiga, Japan). Hybridization, washing, and detection of signals were performed essentially as described previously [32].
GUS staining, DAB staining, and H2O2 measurement
To examine the tissue-specific expression patterns of PPR287, an approximately 1.5-kb fragment of the genomic DNA harboring the putative promoter of PPR287 was cloned into the SphI/BamHI site in front of a GUS reporter gene in pBI121 vector using the primers listed in Additional file 8, and the resulting PPR287PRO::GUS construct was introduced into Arabidopsis by means of vacuum infiltration using Agrobacteruim tumefaciens GV3101 [39]. The transgenic plants were stained in a 0.5 M potassium phosphate buffer (pH 7.0) solution containing 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 10 mM EDTA, 1 mg/ml 5-bromo-4-chloro-3-indole-β-D-glucuronide, 20% methanol, and 0.05% Triton X-100 at 37 °C for 24 h in the dark. After washing the samples with ethanol, GUS images were observed using a Zeiss Axioplan microscope (Carl Zeiss, Inc.). In situ detection of H2O2 was carried out by 3,3′-diaminobenzidine (DAB) staining as previously described [44]. Briefly, 2-week-old leaves were immersed in a solution (0.1% DAB, 0.01 M Na2HPO4, pH 3.8, 0.05% Tween-20), vacuum-infiltrated for 10 min, and incubated overnight at room temperature in the dark. Photographs were taken after bleaching out chlorophylls in an 80% ethanol solution. The level of H2O2 was measured as previously described [45]. Briefly, approximately 200 mg of tissue samples were treated with 0.1% trichloroacetic acid at 4 °C, and the extract was mixed with 100 mM potassium phosphate buffer (pH 7.0) and 1 M KI solution. The reaction mixture was placed in the dark at room temperature for 1 h, and the H2O2 content was determined by measuring the absorbance at 410 nm.
Statistical analysis
The differences in growth parameters, chlorophyll contents, and photosynthetic activity between the wild type, mutants, and complementation lines were compared by t test (p ≤ 0.05) using the SigmaPlot 10 program (Systat Software, Inc., San Jose, CA, USA).
Availability of data and materials
All data can be found within the manuscript and additional files. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- amiRNA:
-
artificial microRNA
- Com:
-
Complementation line
- DAB:
-
3,3′-diaminobenzidine
- Fv/Fm:
-
quantum yield of photosystem II
- GFP:
-
Green fluorescent protein
- H2O2 :
-
Hydrogen peroxide
- KD:
-
Knockdown
- MS:
-
Murashige and Skoog
- NO:
-
Nitric oxide
- O2 − :
-
superoxide
- PPR:
-
Pentatricopeptide repeat
- RBP:
-
RNA-binding protein
- ROS:
-
Reactive oxygen species
References
Abdallah F, Salamini F, Leister D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000;5:141–2.
Leister D. Chloroplast research in the genomic age. Trends Genet. 2003;19:47–56.
Leister D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005;21:655–63.
Surpin M, Larkin RM, Chory J. Signal transduction between the chloroplast and the nucleus. Plant Cell. 2002;14:S327–38.
Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–59.
Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr Opin Plant Biol. 2007;10:600–6.
Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–95.
Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–8.
Marin-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth Res. 2007;94:359–74.
del Campo EM. Post-transcriptional control of chloroplast gene expression. Gene Regul Syst Biol. 2009;3:31.
Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;6:125–55.
Jacobs J, Kück U. Function of chloroplast RNA-binding proteins. Cell Mol Life Sci. 2011;68:737–48.
Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53.
de Souza A, Wang JZ, Dehesh K. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol. 2017;68:85–108.
Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–103.
Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–70.
Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2015;65:415–42.
O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25:1120–8.
Filipovska A, Rackham O. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol BioSyst. 2012;8:699–708.
Gutmann B, Royan S, Small I. Protein complexes implicated in RNA editing in plant organelles. Mol Plant. 2017;10:1255–7.
Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–30.
Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–804.
Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–52.
Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci U S A. 2011;108:415–20.
Zoschke R, Watkins KP, Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25:2265–75.
Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–95.
Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem. 2003;270:4070–81.
Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–48.
Yamazaki H, Tasaka M, Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004;38:152–63.
Cai WH, Okuda K, Peng LW, Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011;67:318–27.
Haili N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, des Francs-Small CC, Vrielynck N, Mireau H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013;41:6650–63.
Lee K, Han JH, Park YI. Colas des Francs-Small C, Small I, Kang H. the mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytol. 2017;215:202–16.
Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, Castleden I, Song Y, Song B, Huang J, Liu X, Xu X, Lim BL, Bond CS, Yiu SM, Small I. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016;85:532–47.
Liu X, Yu F, Rodermel S. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for ftsH-mediated chloroplast biogenesis. Plant Physiol. 2010;154:1588–601.
Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, Howell KA, Huang J. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016;85:607–21.
Zoschke R, Watkins KP, Miranda RG, Barkan A. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016;85:594–606.
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–33.
Kim WY, Jung HJ, Kwak KJ, Kim MK, Oh SH, Han YS, Kang H. The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell. 2010;22:3951–62.
Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–66.
Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–82.
Gu L, Xu T, Lee K, Lee KH, Kang H. A chloroplast-localized DEAD-box RNA helicaseAtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol Biochem. 2014;82:309–18.
Han JH, Lee K, Lee KH, Jung S, Jeon Y, Pai HS, Kang H. A nuclear-encoded chloroplast-targeted S1 RNA-binding domain protein affects chloroplast rRNA processing and is crucial for the normal growth of Arabidopsis thaliana. Plant J. 2015;83:277–89.
Lee K, Lee HJ, Kim DH, Jeon Y, Pai HS, Kang H. A nuclear-encoded chloroplast protein harboring a single CRM domain plays an important role in the Arabidopsis growth and stress response. BMC Plant Biol. 2014;14:98.
Ramel F, Sulmon C, Bogard M, Couee I, Gouesbet G. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol. 2009;9:28.
Kim Y-O, Bae H-J, Cho E, Kang H. Exogenous glutathione enhances mercury tolerance by inhibiting mercury entry into plant cells. Front Plant Sci. 2017;8:683.
Acknowledgements
We thank The Arabidopsis Biological Resource Center for providing the T-DNA mutant seeds.
Funding
This work was financially supported from the Next-Generation BioGreen21 Program (PJ01314701), Rural Development Administration, Republic of Korea, and from the Mid-career Researcher Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2016R1A2B4009172), Republic of Korea. The funding agencies provided funding to the research projects, but played no role in the design of the study, collection, analysis, the interpretation of data and in the writing of this manuscript. These were the sole responsibilities of the authors.
Author information
Authors and Affiliations
Contributions
HK designed the experiments; KL, SJP, JHH, and YJ conducted most of research and analyzed the data together with HK and HSP; KL and HK contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Cellular localization of the GFP only protein. GFP signals from the GFP-expressing transgenic Arabidopsis plants were observed using a confocal microscope. Red signals indicate chloroplast auto-fluorescence. Bar = 10 μm. (PDF 52 kb)
Additional file 2:
Tissue-specific expression patterns of PPR287. GUS activity in (a) 5-day-old seedling, (b) 14-day-old seedling, (c) 30-day-old leaf, (d) 40-day-old stem, (e) and (f) flowers, (g) stigma, and (h) siliques. Bar = 1 cm. (PDF 49 kb)
Additional file 3:
Development-defect phenotypes of artificial microRNA-mediated PPR287 knockdown mutants. (a) Schematic representation of the amiRNA and its target sequence. (b) Downregulation of PPR287 in each knockdown mutant line was determined by RT-PCR and real-time PCR. (c) Phenotypes of knockdown mutants on MS media at 7 days. Bar = 1 cm. (PDF 120 kb)
Additional file 4:
PPR287 plays a role in plant growth and development. (a) Bolting time, (b) leaf number at bolting, (c) plant height, and (d) seed yield of the Col-0, ppr287 mutant, knockdown mutant (KD1), and complementation line (Com1). Values are mean ± SE of three independent experiments (n = 5), and statistically different values are indicated by asterisks (P ≤ 0.05). (e) Growth and leaf morphology at 27 days. Bar = 1 cm. (PDF 161 kb)
Additional file 5:
Abnormal chloroplast structures in ppr287 mutants. The Col-0, ppr287 mutant, knockdown mutant (KD1), and complementation line (Com1) were grown on MS medium, and chloroplast structures in the leaves of 3-week-old plants were observed using a transmission electron microscope. Bar = 1 μm. (PDF 177 kb)
Additional file 6:
Splicing patterns of intron-containing chloroplast genes. (a) Total RNA was extracted from 2-week-old Col-0 (W), ppr287 mutant (K), knockdown mutant (KD1), and complementation line (Com1), and transcript levels of each gene were analyzed by RT-PCR. (b) The rations of un-spliced (precursor) to spliced (mature) transcripts between mutant and wild-type were determined by real-time RT-PCR. Data are mean ± SE of three independent biological replicates. (PDF 81 kb)
Additional file 7:
Expression levels of chloroplast genes. Total RNA was extracted from 2-week-old Col-0, ppr287 mutant, and knockdown mutant (KD1), and transcript levels of chloroplast genes were analyzed by real-time RT-PCR. Data are mean ± SE of three independent biological replicates (PDF 28 kb)
Additional file 8:
Gene-specific primer used in RT-PCR experiments and vector construction (PDF 114 kb)
Additional file 9:
Gene-specific primers used for the analysis of splicing efficiency by real-time RT-PCR. (PDF 143 kb)
Additional file 10:
Gene-specific primers used for the analysis of expression levels of chloroplast genes by quantitative RT-PCR. (PDF 234 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lee, K., Park, S.J., Han, J.H. et al. A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development. BMC Plant Biol 19, 244 (2019). https://doi.org/10.1186/s12870-019-1857-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1857-0