Abstract
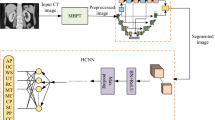
Renal segmentation is one of the most fundamental and challenging task in computer aided diagnosis systems. In order to overcome the shortcomings of automatic kidney segmentation based on deep network for abdominal CT images, a two-stage semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-flow transformation is proposed in paper, which is divided into two stages: image retrieval and semantic segmentation. To facilitate the image retrieval, a metric learning-based approach is firstly adopted to construct a deep convolutional neural network structure using SCNN and ResNet network to extract image features and minimize the impact of interference factors on features, so as to obtain the ability to represent the abdominal CT scan image with the same angle under different imaging conditions. And then, SIFT Flow transformation is introduced, which adopts MRF to fuse label information, priori spatial information and smoothing information to establish the dense matching relationship of pixels so that the semantics can be transferred from the known image to the target image so as to obtain the semantic segmentation result of kidney and space-occupying lesion area. In order to validate effectiveness and efficiency of our proposed method, we conduct experiments on self-establish CT dataset, focus on kidney organ and most of which have tumors inside of the kidney, and abnormal deformed shape of kidney. The experimental results qualitatively and quantitatively show that the accuracy of kidney segmentation is greatly improved, and the key information of the proportioned tumor occupying a small area of the image are exhibited a good segmentation results. In addition, our algorithm has also achieved ideal results in the clinical verification, which is suitable for intelligent medicine equipment applications.








Similar content being viewed by others

References
Hu, P., Wu, F., Peng, J., Bao, Y., Chen, F., and Kong, D., Automatic abdominal multiorgan segmentation using deep convolutional neural network and time-implicit level sets. Int. J. Comput. Assist. Radiol. Surg., 2016a. https://doi.org/10.1007/s11548-016-1501-5.
Li, W., Jia, F., and Hu, Q., Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J. Comput. Commun. 3(11):146–151, 2015.
Vivanti, R., Ephrat, A., Joskowicz, L., Karaaslan, O., Lev-Cohain, N., and Sosna, J., Automatic liver tumor segmentation in follow-up CT studies using convolutional neural networks. Proc. Patch-Based Methods Med. Image Process. Workshop, MICCAI’2015, 54–61, 2015.
Ben-Cohen, A., Diamant, I., Klang, E., Amitai, M., and Greenspan, H., Deep learning and data labeling for medical applications. In: Proceedings of the International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Lect. Notes Comput. Sci. 10008:77–85, 2016. https://doi.org/10.1007/978-3-319-46976-8_9.
Christ, P. F., Elshaer, M. E. A., Ettlinger, F., Tatavarty, S., Bickel, M., Bilic, P., Rempfler, M., Armbruster, M., Hofmann, F., D’Anastasi, M. et al., Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Proceedings of the medical image computing and computer-assisted intervention. Lect. Notes Comput. Sci. 9901:415–423, 2016. https://doi.org/10.1007/978-3-319-46723-8_48.
Dou, Q., Chen, H., **, Y., Yu, L., Qin, J., and Heng, P.-A., 3D deeply supervised network for automatic liver segmentation from CT volumes. IEEE Trans. Biomed. Eng. 64(7):1558–1567, 2016a.
Hu, P., Wu, F., Peng, J., Liang, P., and Kong, D., Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys. Med. Biol. 61:8676–8698, 2016b.
Lu, F., Wu, F., Hu, P., Peng, Z., and Kong, D., Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int. J. Comput. Assist. Radiol. Surg. 12:171–182, 2017.
Lu, X., Xu, D., and Liu, D., Robust 3d organ localization with dual learning architectures and fusion. In: Proceedings of the deep learning in medical image analysis (DLMIA). Lect. Notes Comput. Sci. 10008:12–20, 2016.
Ravishankar, H., Sudhakar, P., Venkataramani, R., Thiruvenkadam, S., Annangi, P., Babu, N., and Vaidya, V., Understanding the mechanisms of deep transfer learning for medical images. In: Proceedings of the Deep Learning in Medical Image Analysis (DLMIA). Lect. Notes Comput. Sci. 10008:188–196, 2016b.
Thong, W., Kadoury, S., Piché, N., and Pal, C.J., Convolutional networks for kidney segmentation in contrast-enhanced CT scans. Comput. Methods Biomech. Biomed. Eng. Imag. Vis. 1–6, 2016..
Farag, A., Lu, L., Roth, H.R., Liu, J., Turkbey, E., and Summers, R.M., A bottom-up approach for pancreas segmentation using cascaded superpixels and (deep) image patch labeling. arxiv:1505.06236, 2015.
Roth, H. R., Lu, L., Farag, A., Shin, H.-C., Liu, J., Turkbey, E. B., and Summers, R. M., DeepOrgan: Multi-level deep convolutional networks for automated pancreas segmentation. In: Proceedings of the medical image computing and ComputerAssisted intervention. Lect. Notes Comput. Sci. 9349:556–564, 2015b.
Cai, J., Lu, L., Zhang, Z., **ng, F., Yang, L., and Yin, Q., Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. Proc. Med. Image Comput. Computer-Assist. Interven. Lect. Notes Comput. Sci. 9901:442–450, 2016a.
Roth, H. R., Lu, L., Farag, A., Sohn, A., and Summers, R. M., S holistically-nested networks for automated pancreas segmeings of the medical image computing and computer-Assi. Lect. Notes Comput. Sci. 9901:451–459, 2016a.
Tajbakhsh, N., Gurudu, S. R., and Liang, J., A comprehensive computer-aided polyp detection system for colonoscopy videos. Proc. Inform. Process. Med. Imag. Lect. Notes Comput. Sci. 9123:327–338, 2015b. https://doi.org/10.1007/978-3-319-19992-4_25.
Liu, J., Wang, D., Wei, Z., Lu, L., Kim, L., Turkbey, E., and Summers, R.M., .Colitis detection on computed tomography using regional convolutional neural networks. Proc. IEEE Int. Symp. Biomed. Imag. 863–866, 2016a.
Nappi, J.J., Hironaka, T., Regge, D., and Yoshida, H., .Deep transfer learning of virtual endoluminal views for the detection of polyps in CT colonography. Proc. Med. Imag. 97852B, 2016.
Tachibana, R., Näppi, J. J., Hironaka, T., Kim, S. H., and Yoshida, H., Deep learning for electronic cleansing in dual-energy ct colonography. Proc. SPIE Med. Imag. 9785:97851M, 2016.
Zhang, R., Zheng, Y., Mak, T. W. C., Yu, R., Wong, S. H., Lau, J. Y. W., and Poon, C. C. Y., Automatic detection and classification of colorectal polyps by transferring lowlevel CNN features from nonmedical domain. IEEE J. Biomed. Health Inf. 21:41–47, 2017.
Liao, S., Gao, Y., Oto, A., and Shen, D., Representation learning: A unified deep learning framework for automatic prostate mr segmentation. In: Proceedings of the medical image computing and computer-assisted intervention. Lect. Notes Comput. Sci. 8150:254–261, 2013.
Cheng, J.-Z., Ni, D., Chou, Y.-H., Qin, J., Tiu, C.-M., Chang, Y.-C., Huang, C.-S., Shen, D., and Chen, C.-M., Computer-aided diagnosis with deep learning architecture: Applications to breast lesions in US images and pulmonary nodules in CT scans. Nat. Sci. Rep. 6:24454, 2016a.
Guo, Y., Gao, Y., and Shen, D., Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans. Med. Imag. 35(4):1077–1089, 2016.
Milletari, F., Navab, N., and Ahmadi, S.-A., V-Net: fully convolutional neural networks for volumetric medical image segmentation. arxiv:1606.04797, 2016b.
Yu, L., Yang, X., Chen, H., Qin, J., and Heng, P.A., Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images. Proc. Thirty-First AAAI Conf. Artif. Intell., 2017c.
Azizi, S., Imani, F., Ghavidel, S., Tahmasebi, A., Kwak, J. T., Xu, S., Turkbey, B., Choyke, P., Pinto, P., Wood, B., Mousavi, P., and Abolmaesumi, P., Detection of prostate cancer using temporal sequences of ultrasound data: A large clinical feasibility study. Int. J. Comput. Assist. Radiol. Surg. 11(6):947–956, 2016.
Shah, A., Conjeti, S., Navab, N., and Katouzian, A., Deeply learnt hashing forests for content based image retrieval in prostate MR images. Proc. SPIE Med. Imag. 9784:978414, 2016.
Zhu, Y., Wang, L., Liu, M., Qian, C., Yousuf, A., Oto, A., and Shen, D., MRI based prostate cancer detection with high-level representation and hierarchical classification. Med. Phys. 44(3):1028–1039, 2017.
Cha, K. H., Hadjiiski, L. M., Samala, R. K., Chan, H.-P., Cohan, R. H., Caoili, E. M., Paramagul, C., Alva, A., and Weizer, A. Z., Bladder cancer segmentation in CT for treatment response assessment: Application of deep-learning convolution neural network-a pilot study. Tomography 2:421–429, 2016.
Acknowledgements
This paper is supported by the Jiangsu Committee of Health on the subject (No. H2018071).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
We declare that we have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
**a, Kj., Yin, Hs. & Zhang, Yd. Deep Semantic Segmentation of Kidney and Space-Occupying Lesion Area Based on SCNN and ResNet Models Combined with SIFT-Flow Algorithm. J Med Syst 43, 2 (2019). https://doi.org/10.1007/s10916-018-1116-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-1116-1