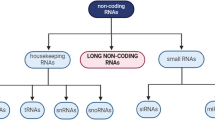
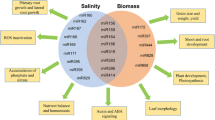
Abstract
Millets are a group of annual, panicoid grasses with C4 mode of photosynthesis. With the ease of their Arabidopsis-like lifecycle, tremendous stress-tolerance capacity, and huge nutritional value, millets can be the genetic and nutritional epitome for worldwide food security as well as for the development of C4 model plant system. Recent sophistications in “omics” approaches may enlighten the path for improvement of the existing genetic resources to sustain over the ongoing environmental dynamics and to incorporate these climate-resilient genetic elements within the established food crops. With the advent of different omics-perspective, long non-coding RNAs (lncRNAs) get huge attention particularly in the prospect for plants’ stress response. The aim of this chapter is to discuss the present state of knowledge about how small millets can be established as the climate-sustainable model system, and how lncRNAs involve in abiotic stress management. It also discusses the utilization of several bioinformatics pipelines to help in unravelling the molecular mystery behind it.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Anuradha N, Satyavathi CT, Bharadwaj C, Nepolean T, Sankar SM, Singh SP, Meena MC, Singhal T, Srivastava RK (2017) Deciphering genomic regions for high grain iron and zinc content using association map** in pearl millet. Front Plant Sci 8:412
Bai Y, Dai X, Harrison AP, Chen M (2015) RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Brief Funct Genomics 14(2):91–101
Bandyopadhyay T, Jaiswal V, Prasad M (2017) Nutrition potential of foxtail millet in comparison to other millets and major cereals. In: The foxtail millet genome. Springer, Cham, pp 123–135
Bhatia G, Singh A, Verma D, Sharma S, Singh K (2020) Genome-wide investigation of regulatory roles of lncRNAs in response to heat and drought stress in Brassica juncea (Indian mustard). Environ Exp Bot 171:103922
Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM (2009) The genetic architecture of maize flowering time. Science 325(5941):714–718
Calixto CP, Tzioutziou NA, James AB, Hornyik C, Guo W, Zhang R, Nimmo HG, Brown JW (2019) Cold-dependent expression and alternative splicing of Arabidopsis long non-coding RNAs. Front Plant Sci 10:235
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44(4):667–678
Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13(21):5099–5112
Deng P, Liu S, Nie X, Weining S, Wu L (2018) Conservation analysis of long non-coding RNAs in plants. Sci China Life Sci 61(2):190–198
Dwivedi SL, Upadhyaya HD, Senthilvel S, Hash CT, Fukunaga K, Diao X, Santra D, Baltensperge D, Prasad M (2012) Millets: genetic and genomic resources. In: Janick J (ed) Plant breeding reviews. Springer, New York. https://doi.org/10.1002/9781118100509.ch5
Fukuda M, Nishida S, Kakei Y, Shimada Y, Fujiwara T (2019) Genome-wide analysis of long intergenic noncoding RNAs responding to low-nutrient conditions in Arabidopsis thaliana: possible involvement of trans-acting siRNA3 in response to low nitrogen. Plant Cell Physiol 60(9):1961–1973
Garg R, Verma M, Agrawal S, Shankar R, Majee M, Jain M (2014) Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res 21(1):69–84
Hatakeyama M, Aluri S, Balachadran MT, Sivarajan SR, Patrignani A, Grüter S, Poveda L, Shimizu-Inatsugi R, Baeten J, Francoijs KJ, Nataraja KN (2018) Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res 25(1):39–47
Jaiswal V, Bandyopadhyay T, Gahlaut V, Gupta S, Dhaka A, Ramchiary N, Prasad M (2019) Genome-wide association study (GWAS) delineates genomic loci for ten nutritional elements in foxtail millet (Setaria italica L.). J Cereal Sci 85:48–55
Jayakodi M, Madheswaran M, Adhimoolam K, Perumal S, Manickam D, Kandasamy T, Yang TJ, Natesan S (2019) Transcriptomes of Indian barnyard millet and barnyard grass reveal putative genes involved in drought adaptation and micronutrient accumulation. Acta Physiol Plant 41(5):1–11
Jha UC, Nayyar H, Jha R, Khurshid M, Zhou M, Mantri N, Siddique KH (2020) Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol 20(1):1–20
Jordan DR, Hunt CH, Cruickshank AW, Borrell AK, Henzell RG (2012) The relationship between the stay-green trait and grain yield in elite sorghum hybrids grown in a range of environments. Crop Sci 52(3):1153–1161
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106(28):11667–11672
Kim DH, ** Y, Sung S (2017) Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet 13(7):e1006939
Kogenaru S, Yan Q, Guo Y, Wang N (2012) RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 13(1):1–16
Kumar S, Hash CT, Nepolean T, Mahendrakar MD, Satyavathi CT, Singh G, Rathore A, Yadav RS, Gupta R, Srivastava RK (2018) Map** grain iron and zinc content quantitative trait loci in an Iniadi-derived immortal population of pearl millet. Genes 9(5):248
Lata CH (2015) Advances in omics for enhancing abiotic stress tolerance in millets. Proc Indian Natl Sci Acad 81:397–417
Lata C, Gupta S, Prasad M (2013) Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 33(3):328–343
Liao Q, Liu C, Yuan X, Kang S, Miao R, **ao H et al (2011) Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res 39(9):3864–3878
Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L et al (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24(11):4333–4345
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12(2):e1005767
Ma P, Zhang X, Luo B, Chen Z, He X, Zhang H, Li B, Liu D, Wu L, Gao S, Gao D (2021) Transcriptomic and genome-wide association study reveal long noncoding RNAs responding to nitrogen deficiency in maize. BMC Plant Biol 21(1):1–19
Mehta PA, Sivaprakash K, Parani M, Venkataraman G, Parida AK (2005) Generation and analysis of expressed sequence tags from the salt-tolerant mangrove species Avicennia marina (Forsk) Vierh. Theor Appl Genet 110(3):416–424
Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy AR, Sopory SK, Reddy MK (2007) Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol Biol 64(6):713–732
Moran VA, Niland CN, Khalil AM (2012a) Co-immunoprecipitation of long noncoding RNAs. In: Genomic imprinting. Humana Press, Totowa, pp 219–228
Moran VA, Perera RJ, Khalil AM (2012b) Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40(14):6391–6400
Ochogavía A, Galla G, Seijo JG, González AM, Bellucci M, Pupilli F, Barcaccia G, Albertini E, Pessino S (2018) Structure, target-specificity and expression of PN_LNC_N13, a long non-coding RNA differentially expressed in apomictic and sexual Paspalum notatum. Plant Mol Biol 96(1):53–67
Peng R, Zhang B (2020) Foxtail millet: a new model for C4 plants. Trends Plant Sci 26(3):199–201
Provart NJ, Alonso J, Assmann SM, Bergmann D, Brady SM, Brkljacic J et al (2016) 50 years of Arabidopsis research: highlights and future directions. New Phytol 209(3):921–944
Qi X, **e S, Liu Y, Yi F, Yu J (2013) Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol 83(4–5):459–473
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Sage RF (2004) The evolution of C4 photosynthesis. New Phytol 161(2):341–370
Salleh FM, Evans K, Goodall B, Machin H, Mowla SB, Mur LA, Runions J, Theodoulou FL, Foyer CH, Rogers HJ (2012) A novel function for a redox‐related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ 35(2):418–429
Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE (2011) The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci 108(51):20497–20502
Sun M, Huang D, Zhang A, Khan I, Yan H, Wang X, Zhang X, Zhang J, Huang L (2020) Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol 20(1):1–15
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462(7274):799–802
Unver T, Tombuloglu H (2020) Barley long non-coding RNAs (lncRNA) responsive to excess boron. Genomics 112(2):1947–1955
Urquiaga MC, Thiebaut F, Hemerly AS, Ferreira PC (2021) From trash to luxury: the potential role of plant LncRNA in DNA methylation during abiotic stress. Front Plant Sci 11:2130
Varshney RK, Terauchi R, McCouch SR (2014) Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol 12(6):e1001883
Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH (2015) Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lnc RNA s associated with agriculture traits. Plant J 84(2):404–416
Wang P, Dai L, Ai J, Wang Y, Ren F (2019) Identification and functional prediction of cold-related long non-coding RNA (lncRNA) in grapevine. Sci Rep 9(1):1–15
Wang T, Song H, Wei Y, Li P, Hu N, Liu J et al (2020) High throughput deep sequencing elucidates the important role of lncRNAs in foxtail millet response to herbicides. Genomics 112(6):4463–4473
Waseem M, Liu Y, **a R (2021) Long non-coding RNAs, the dark matter: an emerging regulatory component in plants. Int J Mol Sci 22(1):86
Werner BT, Gaffar FY, Schuemann J, Biedenkopf D, Koch AM (2020) RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front Plant Sci 11:476
Wu L, Liu S, Qi H, Cai H, Xu M (2020) Research progress on plant long non-coding RNA. Plan Theory 9(4):408
Wunderlich M, Groß-Hardt R, Schöffl F (2014) Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 85(6):541–550
Yang Z, Zhang H, Li X, Shen H, Gao J, Hou S, Zhang B, Mayes S, Bennett M, Ma J, Wu C (2020) A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat Plants 6(9):1167–1178
Yoon JH, Abdelmohsen K, Gorospe M (2014) Functional interactions among microRNAs and long noncoding RNAs. In: Seminars in cell & developmental biology, vol 34. Academic Press, Cambridge, pp 9–14
Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP (2012) Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol 4(4):427–442
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association map** in maize. Genetics 178(1):539–551
Zhang D, Zhang B (2020) SpRY: engineered CRISPR/Cas9 harnesses new genome-editing power. Trends Genet 36(8):546–548
Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, **e M, Zeng P, Yue Z, Wang W, Tao Y (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30(6):549–554
Zhang C, Tang G, Peng X, Sun F, Liu S, ** Y (2018) Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses. BMC Plant Biol 18(1):1–15
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40(6):939–953
Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P, Durand JL (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci 114(35):9326–9331
Zhao Z, Liu D, Cui Y, Li S, Liang D, Sun D, Wang J, Liu Z (2020) Genome-wide identification and characterization of long non-coding RNAs related to grain yield in foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 21(1):1–13
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Mal, N., Mal, C. (2022). Abiotic Stress Tolerant Small Millet Plant Growth Regulation by Long Non-coding RNAs: An Omics Perspective. In: Pudake, R.N., Solanke, A.U., Sevanthi, A.M., Rajendrakumar, P. (eds) Omics of Climate Resilient Small Millets. Springer, Singapore. https://doi.org/10.1007/978-981-19-3907-5_15
Download citation
DOI: https://doi.org/10.1007/978-981-19-3907-5_15
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-3906-8
Online ISBN: 978-981-19-3907-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)