Abstract
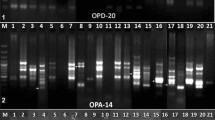
Bulgaria was the secondary gene pool for many crops, and one of the first was pepper. However, during the political transforming and economic crises, the lands for growing pepper (Capsicum spp.) were reduced, and thereafter, the genetic diversity was lost. With pepper, Bulgaria still has priority providing on European scale valuable pepper germplasm, and this priority should be evaluated and preserved. We present our efforts to characterize pepper accessions using RAPD as well as the retroelement-based Inter-SINE Amplified Polymorphism (ISAP) method initially developed for potatoes. Several short interspersed nuclear element (SINE) families were active within the common ancestor of potato and pepper. We studied the degree of polymorphisms in a collection of 73 pepper genotypes, divided into six groups, using ISAP with primers derived from seven Solanaceae SINE families as well as two subfamilies. Two primer pairs from the families SolS-II and SolS-V generated the most fragments and most informative banding patterns. These SINE-based ISAP reactions are best suited for identifying species of the Capsicum genus. The most polymorphic profiles within all studied were generated by C. baccatum accessions. In contrast, intraspecific application of the SINE-based markers yielded a high percentage of conserved ISAP fragments. From a total of 56 C. annuum accessions, only three of them with two different profiles were identified. Our results demonstrate that potato-based SolS-SINE primers can be adapted for molecular genoty** in peppers. The low intraspecies polymorphism generated by ISAP forced us to investigate RAPD as an alternative low-cost genoty** approach. RAPD was successfully applied on a group of mutant lines and corresponding source lines, carrying valuable breeding traits. Despite the low polymorphic levels, we have identified four RAPD primers, capable to discriminate among several genotypes.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- BSA:
-
Bovin serum albumin
- CTAB:
-
Hexadecyltrimethylammonium bromide [(C16H33)N(CH3)3]Br
- EDTA:
-
Ethylenediaminetetraacetic acid [CH2N(CH2CO2H)2]2
- M:
-
Multiplex reaction
- Sol:
-
Solanaceae
References
Adetula, O. (2005). Genetic diversity of capsicum using random amplified polymorphic DNAs. African Journal of Biotechnology, 5, 120–122.
Ahn, Y., Tripathi, S., Kim, J., Cho, Y., Lee, H., Kim, D., Woo, J., & Yoon, M. (2014). Microsatellite marker information from high-throughput next-generation sequence data of Capsicum annuum varieties Mandarin and Blackcluster. Scientia Horticulturae, 170, 123–130.
Alzohairy, A. M., Gyulai, G., Mustafa, M., Edris, S., Sabir, J., Jansen, R. K., & Bahieldin, A. (2015). Retrotransposon-based plant DNA barcoding. In M. Ajmal Ali, G. Gyulai, & F. A. Hemaid (Eds.), Plant DNA barcoding and Phylogenetics (pp. 1–14). Lambert Academic Publishing.
Arnedo-Andrés, M., Gil-Ortega, R., Luis-Arteaga, M., & Hormaza, J. (2002). Development of RAPD and SCAR markers linked to the Pvr4 locus for resistance to PVY in pepper (Capsicum annuum L.). Theoretical and Applied Genetics, 105, 1067–1074.
Bahurupe, J. V., Sakhare, S. B., Kulwal, P. L., Akhare, A. A., & Pawar, B. D. (2013). Genetic diversity analysis in chilli (Capsicum annuum L.) using RAPD markers. The Bioscan, An International Quarterly Journal of Life Sciences, 8, 915–918.
Ballester, J., & Carmen de Vicente, M. (1998). Determination of F1 hybrid seed purity in pepper using PCR-based markers. Euphytica, 103, 223–226.
Baoxi, Z., Sanwen, H., Guimei, Y., & Jiazhen, G. (2000). Two RAPD markers linked to a major fertility restorer gene in pepper. Euphytica, 113, 155–161.
Bello-Bello, J., Iglesias-Andreu, L., Avilés-Viñas, S., Gómez-Uc, E., Canto-Flick, A., & Santana-Buzzy, N. (2014). Somaclonal variation in habanero pepper (Capsicum chinense Jacq.) as assessed ISSR molecular markers. Hort Science, 49, 481–485.
Biedler, J., & Tu, Z. (2003). Non-LTR retrotransposons in the African malaria mosquito, Anopheles gambiae: Unprecedented diversity and evidence of recent activity. Molecular Biology аnd Evolution, 20, 1811–1825.
Cost, G. J., & Boeke, J. D. (1998). Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry, 37, 18081–18093.
Deragon, J., & Zhang, X. (2006). Short Interspersed Elements (SINEs) in plants: Origin, classification, and use as phylogenetic markers. Systematic Biology, 55, 949–956.
Eickbush, T. H., & Malik, H. S. (2002). In N. L. Craig, R. Craigie, M. Gellert, & A. M. Lambowitz (Eds.), Mobile DNA II (pp. 1111–1146). ASM, Herndon.
Ferguson, A., Zhao, D., & Jiang, N. (2013). Selective acquisition and retention of genomic sequences by Pack-Mutator-like elements based on guanine-cytosine content and the breadth of expression. Plant Physiology, 163, 1419–1432.
Feschotte, C., Jiang, N., & Wessler, S. (2002). Plant transposable elements: Where genetics meets genomics. Macmillan Magazines, 3, 329–341.
Geleta, L. F., Labuschagne, M. Y., & Viljoen, C. D. (2005). Genetic variability in pepper (Capsicum annuum L.) estimated by morphological data and amplified fragment length polymorphism markers. Biodiversity and Conservation, 14, 2361–2375.
Gozukirmizi, N., Yilmaz, S., Marakli, S., & Temel, A. (2015). Retrotransposon-based molecular markers; tools for variation analysis in plants. Applications of molecular markers in plant genome analysis and breeding, Eds. Ksenija Taški-Ajduković Research Signpost.
Hazarika, R., & Neog, B. (2014). Investigation of intraspecific diversity in Capsicum chinense using morphological and molecular markers. Indian Journal of Genetics and Plant Breeding, 74, 392–395.
Hedges, D. J., & Batzer, M. A. (2005). From the margins of the genome: Mobile elements shape primate evolution. BioEssays, 27, 785–794.
Ibarra-Torresa, P., Valadez-Moctezumab, E., Pérez-Grajalesb, M., Rodríguez-Camposc, J., & Jaramillo-Floresa, M. (2015). Inter- and intraspecific differentiation of Capsicum annuum and Capsicum pubescens using ISSR and SSR markers. Scientia Horticulturae, 181, 137–146.
Ilbi, H. (2003). RAPD markers assisted varietal identification and genetic purity test in pepper, Capsicum annuum. Scientia Horticulturae, 97, 211–218.
Jang, I., Moon, J. H., Yoon, J. H., Yang, T. J., Kim, Y. J., & Park, H. G. (2004). Application of RAPD and SCAR markers for purity testing of F1 hybrid seed in chili pepper (Capsicum annuum). Molecular Cell, 18, 295–299.
Kajikawa, M., Ichiyanagi, K., Tanaka, N., & Okada, N. (2005). Isolation and characterization of active LINE and SINEs from the Eel. Molecular Biology and Evolution, 22, 673–682.
Kalendar, R., & Schulman, A. H. (2006). IRAP and REMAP for retrotransposon-based genoty** and fingerprinting. Nature Protocols, 12478–12484.
Kalendar, R., Vicient, C. M., Peleg, O., Anamthawat-Jonsson, K., Bolshoy, A., & Schulman, A. (2004). Large retrotransposon derivatives: Abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics, 166, 1437–1450.
Kalendar, R., Flavell, A. J., Ellis, T., Sjakste, T., Moisy, C., & Schulman, A. H. (2011). Analysis of plant diversity with retrotransposon-based molecular markers. Heredity (Edinburgh), 106, 520–530.
Kim, S., Park, M., Yeom, S., Kim, Y. M., Lee, J. M., Lee, A. H., Seo, E., Choi, J., Cheong, K., Kim, T. K., Lee, W. G., Oh, K. S., Bae, C., Kim, S. B., Lee, Y. H., Kim, S. Y., Kim, M. S., Kang, C. C., Jo, D. Y., Yang, H. B., Jeong, J. H., Kang, H. W., Kwon, J. K., Shin, C., Lim, Y. J., Park, H. J., Huh, H. J., Kim, S. J., Kim, D. B., Cohen, O., Paran, I., Suh, C. M., Lee, B. S., Kim, K. Y., Shin, Y., Noh, J. S., Park, J., Seo, S. Y., Kwon, Y. S., Kim, A. H., Park, M. J., Kim, J. H., Chio, S. H., Lee, S. M., Yu, Y., Choi, D. Y., Park, S. B., Deynze, A., Ashrafi, H., Hill, T., Kim, T. W., Pai, S. H., Ahn, K. H., Yeam, I., Giovannoni, J. J., Rose, K. J., Sorensen, T. W., Lee, J. S., Kim, W. R., Choi, Y. I., Choi, S. B., Lim, S. J., Lee, H. Y., & Choi, D. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in capsicum species. Nature Genetics, 46, 270–278.
Kumar, A., Pearce, S., McLean, K., Harrison, G., Heslop-Harrison, J. S., Waugh, R., & Flavell, A. J. (1997). The Ty1-copia group of retrotransposons in plants: Genomic organisation, evolution, and use as molecular markers. Genetica, 100(1–3), 205–217. https://doi.org/10.1023/A:1018393931948
Kumar, S., Singh, V., Singh, M., Raia, S., Kumar, S., Rai, S., & Rai, M. (2007). Genetics and distribution of fertility restoration associated RAPD markers in inbreds of pepper (Capsicum annuum L.). Scientia Horticulturae, 111, 197–202.
Lee, H., Ayarpadikannan, S., & Kim, H. (2015). Role of transposable elements in genomic rearrangement, evolution, gene regulation and epigenetics in primates. Genes and Genetic Systems, 90, 245–257.
Li, G., & Quiros, C. F. (2001). Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to map** and gene tagging in Brassica. Theoretical Applied Genetics, 103, 455–461.
Lijun, O., & Xuexiao, Z. (2012). Inter simple sequence repeat analysis of genetic diversity of five cultivated pepper species. African Journal of Biotechnology, 11, 752–757.
Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., McCombie, W. R., Colot, V., & Martienssen, R. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature, 430, 471–476.
Livingstone, K. D., Lackney, K., Blauth, R. J., Van Wijk, R., & Jahn, M. K. (1999). Genome map** in capsicum and evolution of genome structure in Solanaceae. Genetics, 155, 1183–1202.
Min, W. K., Han, J. H., Kang, W. H., Lee, H. R., & Kim, B. D. (2008). Reverse random amplified microsatellite polymorphism reveals enhanced polymorphisms in the 3’ end of simple sequence repeats in the pepper genome. Molecules and Cells, 26, 250–257.
Minamiyama, Y., & Tsuro, M. H. (2006). An SSR-based linkage map of Capsicum annuum. Molecular Breeding, 18, 157–169.
Moodley, V., Ibaba, J., Naidoo, R., & Gubba, A. (2014). Full-genome analyses of a Potato Virus Y (PVY) isolate infecting pepper (Capsicum annuum L.) in the Republic of South Africa. Virus Genes, 49, 466–476.
Moore, J. K., & Haber, E. J. (1996). Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature, 383, 644–646.
Munoz-Lopez, M., & Perez-Garacia, J. (2010). DNA transposons: Nature and applications in genomics. Current Genomics, 11, 115–128.
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8, 4321–4325.
Ogundiwin, E. A., Berke, T. F., Massoudi, M., Black, L. L., Huestis, G., Choi, D., Lee, S., & Prince, J. P. (2005). Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome, 48, 698–711.
Paran, I. (2013). Molecular linkage maps of capsicum. In B. Kang & C. Kole (Eds.), Genetics, genomics and breeding of peppers and eggplants (pp. 40–55). Taylor & Francis Group, 161 p.
Park, M., Jo, S., Kwon, J.-K., & Choi, D. (2011). Comparative analysis of pepper and tomato reveals euchromatin expansion of pepper genome caused by differential accumulation of Ty3/Gypsy-like elements. BMC Genomics, 12, 85.
Poryazov, I., Petkova, V., & Tomlekova, N. (2013). Vegetable production. “DIMI 99” Ltd. 318 p. /Bulgarian/.
Pradeepkumar, T., Karihaloo, J. L., & Archak, S. (2001). Molecular characterization of Piper nigrum L. cultivars using RAPD markers. Current Sciences, 81, 245–248.
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., Cheng, J., Zhao, S., Xu, M., Luo, Y., Yang, Y., Wu, Z., Mao, L., Wu, H., Ling-Hu, C., Zhou, H., Lin, H., González-Morales, S., Trejo-Saavedra, D., Tian, H., Tang, X., Zhao, M., Huang, Z., Zhou, A., Yao, X., Cui, J., Li, W., Chen, Z., Feng, Y., Niu, Y., Bi, S., Yang, X., Li, W., Cai, H., Luo, X., Montes-Hernández, S., Leyva-González, M., **ong, Z., He, X., Bai, L., Tan, S., Tang, X., Liu, D., Liu, J., Zhang, S., Chen, M., Zhang, L., Zhang, L., Zhang, Y., Liao, W., Zhang, Y., Wang, M., Lv, X., Wen, B., Liu, H., Luan, H., Zhang, Y., Yang, S., Wang, X., Xu, J., Li, X., Li, S., Wang, J., Palloix, A., Bosland, P., Li, Y., Krogh, A., Rivera-Bustamante, R., Herrera-Estrella, L., Yin, Y., Yu, J., Hu, K., & Zhang, Z. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceeding of the National Academy of Sciences of the United States of American, 111, 5135–5140.
Ravindran, S. (2012). Barbara McClintock and the discovery of jum** genes. Proceeding of the National Academy of Sciences of the United States of America, 109, 20198–20199.
Ray, D. A. (2007). SINEs of progress: Mobile element applications to molecular ecology. Molecular Ecology, 16, 19–33.
Rodriguez, J. M., Berke, T., Engle, L., & Nienhuis, J. (1999). Variation among and within Capsicum species revealed by RAPD markers. Theoretical Applied Genetics, 99, 147–156.
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., & Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the United States of America, 81, 8014–8018.
Schmidt, T. (1999). LINEs, SINEs and repetitive DNA: Non-LTR retrotransposons in plant genomes. Plant Molecular Biology, 40, 903–910.
Schmidt, T., & Heslop-Harrison, J. S. (1998). Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends in Plant Science, 3, 195–199.
Schulman, A. (2007). A unified classification system for eukaryotic transposable elements. Genetics, 8, 973–982.
Seibt, K. M., Wenke, T., Wollrab, C., Junghans, H., Muders, K., Dehmer, K. J., Diekmann, K., & Schmidt, T. (2012). Development and application of SINE-based markers for genoty** of potato varieties. Theoretical Applied Genetics, 125, 185–196.
Seibt, K. M., Wenke, T., Muders, K., Trubergm, B., & Schmidt, T. (2016). Short interspersed nuclear elements (SINEs) are abundant in Solanaceae and have a family-specific impact on gene structure and genome organization. The Plant Journal, 86, 268–285.
Sundaram, V., Cheng, Y., Ma, Z., Li, D., **ng, X., Edge, P., Snyder, M., & Wang, T. (2014). Wide spread contribution of transposable elements to the innovation of gene regulatory networks. Cold Spring Harbor Laboratory Press, 26, 1–14.
Tam, S. M., Mhiri, C., Vogelaar, A., Kerkveld, M., Pearce, S. R., & Marie-Angèle, G. (2005). Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theoretical and Applied Genetics, 110, 819–831.
Tanksley, S. D., Bernatzky, R., Lapitan, N. L., & Prince, J. P. (1988). Conservation of gene repertoire but not gene order in pepper and tomato. Proceedings of the National Academy of Sciences of the United States of America, 85, 6419–6423.
Thul, S., Darokar, M., Shasany, A., & Khanuja, S. (2012). Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Molecular Biotechnology, 51, 137–147.
Todorova, V., Boteva, H., Masheva, S., Cholakov, T., Kostova, D., Yankova, V., & Dincheva, T. S. (2014). Technologies for field pepper production. In S. Masheva, M. Mihov, V. Todorova, E. Nacheva, V. Yankova, & H. Boteva (Eds.), Technologies for production of vegetable crops and potatoes (pp. 41–66). Bulgarian, first edition, circulation 300, Blakom printing house – Plovdiv. 245 p. /Bulgarian/.
Tomlekova, N., Spasova-Apostolova, V., & Panchev, I. (2016). RAPD analysis of Bulgarian pepper induced mutants. Comptes rendus de l’Académie bulgare des sciences, 69(6), 731–738.
Tomlekova, N., Spasova-Apostolova, V., Nacheva, E., Stoyanova, M., Teneva, A., Petrov, N., & Schmidt, T. (2017a). Genoty** of Bulgarian potato varieties by SINE–based ISAP markers. Comptes rendus de l’Académie bulgare des Sciences, 70, 61–70.
Tomlekova, N. B., White, P. J., Thompson, J. A., Penchev, E., & Nielen, S. (2017b). Mutation affecting fruit colour increases β-carotene concentration in sweet pepper. PLoS One, 12(2), e0172180. https://doi.org/10.1371/journal.pone.0172180
Tomlekova, N., Spasova-Apostolova, V., Pantchev, I., & Sarsu, F. (2021). Mutation associated with orange fruit colour increases concentrations of β-carotene in a sweet pepper variety (Capsicum annuum L.). Food, 10(6), 1225. https://doi.org/10.3390/foods10061225
Toquica, S., Rodríguez, F., Martínez, E., Duque, M., & Tohme, J. (2003). Molecular characterization by AFLPs of capsicum germplasm from the Amazon. Genetic Resources and Crop Evolution, 50, 639–647.
Van de Lagemaat, L. N., Gagnier, L., Medstrand, P., & Mager, D. L. (2005). Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Research, 15, 1243–1249.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., Van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., & Zabeau, M. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research, 23, 4407–4414.
Votava, E., Nabhan, G., & Bosland, P. (2002). Genetic diversity and similarity revealed via molecular analysis among and within an in-situ population and ex situ accessions of chiltepín (Capsicum annuum var. glabriusculum). Conservation Genetics, 3, 123–129.
Wang, T., Zeng, J., Lowe, C. B., Sellers, R. G., Salama, S. R., Yang, M., Burgess, S. M., Brachmann, R. K., & Haussler, D. (2007). Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p 53. Proceedings of the National Academy of Sciences of the United States of America, 104, 18613–18618.
Weiner, A. M., Deininger, P. L., & Efstratiadis, A. (1986). Nonviral retroposons: Genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annual Review of Biochemistry, 55, 631–661.
Welsh, J., & McClelland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research, 18, 7213–7218.
Wenke, T., Döbel, T., Sörensen, T. R., Junghans, H., Weisshaar, B., & Schmidt, T. (2011). Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell, 23, 3117–3128.
Wessler, S. (1998). Transposable elements associated with normal plant genes. Physiologia Plantarum, 103, 581–586.
Wessler, S. (2006). Transposable elements and the evolution of eukaryotic genomes. Proceedings of the National Academy of Sciences of the United States of America, 103, 17600–17601.
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., Flavel, A., Leroy, P., Morgante, M., Panaud, O., Paux, E., Miguel, S., & Schulman, A. H. (2007). A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics, 8, 973–982.
Williams, J., Kubelik, A. R., Livak, K., Rafalski, J. A., & Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531–6535.
Winkfein, R. J., Moir, R. D., Krawetz, S. A., Blanco, J., States, J. C., & Dixon, G. H. (1988). A new family of repetitive, retroposon-like sequences in the genome of the rainbow trout. European Journal of Biochemistry, 176, 255–264.
Wu, F., & Tanksley, S. D. (2010). Chromosomal evolution in the plant family Solanaceae. BMC Genomics, 11, 182.
Wu, F., Eannetta, N. T., Xy, Y., Durrett, R., Mazourek, M., Jahn, M. M., & Tanksley, S. D. (2009). A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theoretical Applied Genetics, 118, 1279–1293.
**ong, Y., & Eickbush, T. H. (1990). Origin and evolution of retroelements based upon their reverse transcriptase sequences. The EMBO Journal, 9, 3353–3362.
Xu, X., Zeng, L., Li, Y., & Wang, H. (2014). Inheritance of resistance to Phythophtora capsici in Capsicum annuum and analysis of relative SRAP markers. Journal of Chemical and Pharmaceutical Research, 6, 1967–1972.
Acknowledgments
The authors acknowledge financial support from the International Atomic Energy Agency, grant numbers BUL/5/016 and RER/5/024. We are grateful for the contribution of Prof. Thomas Schmidt (Dresden University of Technology, Germany) in the development and introduction of the ISAP technique as a means of identifying and studying members of the family Solanaceae.
Funding
This research was funded by International Atomic Energy Agency, Technical Cooperation projects BUL/5/016 and RER/5/024.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Tomlekova, N. et al. (2023). Applicability of ISAP and RAPD Techniques for Capsicum Collection Genoty**. In: Raina, A., Wani, M.R., Laskar, R.A., Tomlekova, N., Khan, S. (eds) Advanced Crop Improvement, Volume 2. Springer, Cham. https://doi.org/10.1007/978-3-031-26669-0_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-26669-0_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-26668-3
Online ISBN: 978-3-031-26669-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)