Abstract
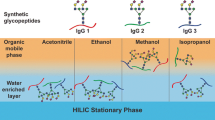
Glycosylation of proteins is an important post-translational modification that plays a role in a wide range of biological processes, including immune response, intercellular signaling, inflammation, and host-pathogen interaction. Abnormal protein glycosylation has been correlated with various diseases. However, the study of protein glycosylation remains challenging due to its low abundance, microheterogeneity of glycosylation sites, and low ionization efficiency. During the past decade, several methods for enrichment and for isolation of glycopeptides from biological samples have been developed and successfully employed in glycoproteomics research. In this chapter, we discuss the sample preparation protocol and the strategies for effectively isolating and enriching glycopeptides from biological samples, using PolyHYDROXYETHYL A as a hydrophilic interaction liquid chromatography (HILIC) enrichment technique.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hart GW, Copeland RJ (2010) Glycomics hits the big time. Cell 143(5):672–676. https://doi.org/10.1016/j.cell.2010.11.008
Tzeng SF, Tsai CH, Chao TK et al (2018) O-Glycosylation-mediated signaling circuit drives metastatic castration-resistant prostate cancer. FASEB J:fj201800687. https://doi.org/10.1096/fj.201800687
de Vreede G, Morrison HA, Houser AM et al (2018) A drosophila tumor suppressor gene prevents tonic TNF signaling through receptor N-glycosylation. Dev Cell 45(5):595–605.e594. https://doi.org/10.1016/j.devcel.2018.05.012
Singh C, Shyanti RK, Singh V et al (2018) Integrin expression and glycosylation patterns regulate cell-matrix adhesion and alter with breast cancer progression. Biochem Biophys Res Commun 499(2):374–380. https://doi.org/10.1016/j.bbrc.2018.03.169
Oyama M, Kariya Y, Kariya Y et al (2018) Biological role of site-specific O-glycosylation in cell adhesion activity and phosphorylation of osteopontin. Biochem J 475(9):1583–1595. https://doi.org/10.1042/bcj20170205
Solá RJ, Griebenow K (2009) Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci 98(4):1223–1245. https://doi.org/10.1002/jps.21504
Desko MM, Gross DA, Kohler JJ (2009) Effects of N-glycosylation on the activity and localization of GlcNAc-6-sulfotransferase 1. Glycobiology 19(10):1068–1077. https://doi.org/10.1093/glycob/cwp092
Sperandio M, Gleissner CA, Ley K (2009) Glycosylation in immune cell trafficking. Immunol Rev 230(1):97–113. https://doi.org/10.1111/j.1600-065X.2009.00795.x
Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473(1):4–8. https://doi.org/10.1016/s0304-4165(99)00165-8
Shental-Bechor D, Levy Y (2008) Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci U S A 105(24):8256–8261. https://doi.org/10.1073/pnas.0801340105
Kizuka Y, Kitazume S, Taniguchi N (2017) N-glycan and alzheimer’s disease. Biochim Biophys Acta Gen Subj 1861(10):2447–2454. https://doi.org/10.1016/j.bbagen.2017.04.012
Van Scherpenzeel M, Willems E, Lefeber DJ (2016) Clinical diagnostics and therapy monitoring in the congenital disorders of glycosylation. Glycoconj J 33(3):345–358. https://doi.org/10.1007/s10719-015-9639-x
Magalhães A, Duarte HO, Reis CA (2021) The role of O-glycosylation in human disease. Mol Asp Med 79:100964. https://doi.org/10.1016/j.mam.2021.100964
Mondello S, Sandner V, Goli M et al (2022) Exploring serum glycome patterns after moderate to severe traumatic brain injury: a prospective pilot study. eClinicalMedicine 50:101494. https://doi.org/10.1016/j.eclinm.2022.101494
Mehta A, Herrera H, Block T (2015) Glycosylation and liver cancer. Adv Cancer Res 126:257–279. https://doi.org/10.1016/bs.acr.2014.11.005
Peng W, Goli M, Mirzaei P et al (2019) Revealing the biological attributes of N-glycan isomers in breast cancer brain metastasis using porous graphitic carbon (PGC) liquid chromatography-tandem mass spectrometry (LC-MS/MS). J Proteome Res 18(10):3731–3740. https://doi.org/10.1021/acs.jproteome.9b00429
Oliveira-Ferrer L, Legler K, Milde-Langosch K (2017) Role of protein glycosylation in cancer metastasis. Semin Cancer Biol 44:141–152. https://doi.org/10.1016/j.semcancer.2017.03.002
Yu A, Zhao J, Peng W et al (2018) Advances in mass spectrometry-based glycoproteomics. Electrophoresis 39(24):3104–3122. https://doi.org/10.1002/elps.201800272
Peng W, Gutierrez Reyes CD, Gautam S et al (2023) MS-based glycomics and glycoproteomics methods enabling isomeric characterization. Mass Spectrom Rev 42(2):577–616. https://doi.org/10.1002/mas.21713
Goli M, Yu A, Cho BG et al (2021) Chapter 8 – LC-MS/MS in glycomics and glycoproteomics analyses. In: El Rassi Z (ed) Carbohydrate analysis by modern liquid phase separation techniques, 2nd edn. Elsevier, Amsterdam, pp 391–441. https://doi.org/10.1016/B978-0-12-821447-3.00005-6
Banazadeh A, Veillon L, Wooding KM et al (2017) Recent advances in mass spectrometric analysis of glycoproteins. Electrophoresis 38(1):162–189. https://doi.org/10.1002/elps.201600357
Gutierrez-Reyes CD, Jiang P, Atashi M et al (2022) Advances in mass spectrometry-based glycoproteomics: an update covering the period 2017-2021. Electrophoresis 43(1–2):370–387. https://doi.org/10.1002/elps.202100188
Cummings RD, Pierce JM (2014) The challenge and promise of glycomics. Chem Biol 21(1):1–15. https://doi.org/10.1016/j.chembiol.2013.12.010
**ao H, Sun F, Suttapitugsakul S et al (2019) Global and site-specific analysis of protein glycosylation in complex biological systems with mass spectrometry. Mass Spectrom Rev 38(4–5):356–379. https://doi.org/10.1002/mas.21586
Gutierrez Reyes CD, Jiang P, Donohoo K et al (2021) Glycomics and glycoproteomics: approaches to address isomeric separation of glycans and glycopeptides. J Sep Sci 44(1):403–425. https://doi.org/10.1002/jssc.202000878
Liu L, Qin H, Ye M (2021) [Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses]. Se Pu 39(10):1045–1054. https://doi.org/10.3724/sp.J.1123.2021.06011
Sun N, Wu H, Chen H et al (2019) Advances in hydrophilic nanomaterials for glycoproteomics. Chem Commun (Camb) 55(70):10359–10375. https://doi.org/10.1039/c9cc04124a
Zhang H, Yi EC, Li XJ et al (2005) High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics 4(2):144–155. https://doi.org/10.1074/mcp.M400090-MCP200
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Goli, M., Jiang, P., Fowowe, M., Hakim, M.A., Mechref, Y. (2024). Hydrophilic Interaction Liquid Chromatography (HILIC) Enrichment of Glycopeptides Using PolyHYDROXYETHYL A. In: Bradfute, S.B. (eds) Recombinant Glycoproteins. Methods in Molecular Biology, vol 2762. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3666-4_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3666-4_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3665-7
Online ISBN: 978-1-0716-3666-4
eBook Packages: Springer Protocols