Abstract
Background
The MYB family is one of the most significant groups of transcription factors in plants. However, several MYBs have been linked to secondary metabolism and are important for determining the color of fruit’s peel and pulp. Despite being a substantial fruit crop in tropical and subtropical areas of the world, wilt-resistant hybrid guava (Psidium guajava × Psidium molle; PGPM) has not yet been the subject of a thorough examination. This study’s goal was to assess the expression of MYB in guava fruit pulp, roots, and seeds to predict its function by in silico analysis of the guava root transcriptome data.
Results
In the current study, we have mined the MYBs family of MYB genes from the transcriptome of the PGPM guava root. We have mined 15 distinct MYB transcription factor genes/transcripts viz MYB3, MYB4, MYB23, MYB86, MYB90, MYB308, MYB5, MYB82, MYB114, MYB6, MYB305, MYB44, MYB51, MYB46, and MYB330. From the analyses, it was found that R2-MYB and R3-MYB domains are conserved in all known guava MYB proteins. The expression of six different MYB TFs was examined using semi-quantitative RT-PCR in “Shweta” pulp (white colour pulp), “Lalit” pulp (red color pulp), “Lalit” root, and “Lalit” seed.
Conclusion
There were 15 MYB family members observed in guava. They were unequally distributed across the chromosomes, most likely as a result of gene duplication. Additionally, the expression patterns of the particular MYBs showed that MYB may be involved in the control of wilt, fruit ripening, seed development, and root development. Our results allow for a more thorough functional characterization of the guava MYB family genes and open the door to additional research into one essential MYB transcription factor family of genes and its involvement in the growth and ripening of guava fruit.
Similar content being viewed by others
Background
Multigene families govern the growth and development of plants. A DNA-binding domain, a nuclear localization signal, a transcription activation domain, and an oligomerization site are the four primary domains that transcription factors typically contain and that play a significant role in the regulation of gene transcription [1]. Through the activation or suppression of the transcriptional process, these four domains cooperate to regulate a wide range of aspects of plant development and growth [2]. Analysis of each transcription factor’s distinct roles is complicated by the fact that multigene families frequently encode transcription factors [1]. The MYB transcription factors of higher plants are more widely distributed across the genome than those of fungi and mammals [3].
MYB transcription factors are made up of two unique regions: an N-terminal conserved MYB DNA binding domain and a C-terminal variable modulator region that regulates the activities of proteins. Plants have a high degree of conservation of the MYB domain, at N-terminus and proteins typically contain one to four repeats viz R1, R2, R3, and R4. Each repetition contains 50–53 amino acids that code for three α-helices, from which the second and third α-helices form the HTH (helix-turn-helix) structure [4]. The third α-helix interacts with the major groove of DNA and forms the transcription factor DNA recognition site [5]. A set of highly conserved tryptophan (W) residues found in the MYB domain is involved in sequence-specific DNA binding [6]. Contrarily, the C-terminal, promoter domain of various MYB proteins is extremely varied, contributing to the wide range of regulatory tasks played by the MYB gene family [7,8,9]. Higher plants often include R2R3-MYB domain proteins as their predominant form [10]. The regulation of primary and secondary metabolism, the management of the cell cycle, and the response to abiotic and biotic stressors are just a few of the phases of plant growth and development where the plant MYB transcription factors have been shown to play a role [11, 12].
The guava (Psidium guajava L.; PG) is a significant fruit crop in tropical and subtropical regions of the world. Guava has 2n = 22 chromosomes and a genomic size of over 450 MB. It belongs to the family Myrtaceae, which comprises about 150 species [13]. Guava is mostly produced in India, Mexico, Pakistan, Taiwan, Thailand, Colombia, and Indonesia, with small-scale plantations also operating in Malaysia, Australia, and South Africa [14]. There are approximately about 400 guava varieties produced all over the world, each with a unique fruit pulp and peel colour. When a fruit reaches maturity, the peel changes from green to yellow or red, and the fruit pulp can range from white to deep pink. This trait varies between cultivars and depends on the climate [15].
Consumers typically choose visually appealing colorful fruits because they have better nutritional characteristics and attractiveness. Genes involved in secondary metabolic pathways, particularly phenylalanine ammonia-lyase (PAL), anthocyanidin synthase (ANS), dihydro-flavonol 4-reductase (DFR), chalcone synthase (CHS), flavanol synthase/flavanone 3-hydroxylase (F3H), UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT), MYB transcription factors, basic helix-loop-helix (bHLH), tryptophan-aspartic acid repeat set c. regulate the color of fruits and vegetables [16,17,18,19,20,21].
Although Psidium guajava L (PG) is a lucrative economic crop it suffers huge losses due to wilt susceptibility. The specific etiology of wilt is unknown, but it has been linked to the pathogens Fusarium oxysporum, Fusarium solani, Rhizoctonia bataticola, Macrophomina phaeseoli, Gliocladium roseum, and Cephalosporium sp. etc. To combat wilt, the ICAR-Central Institute for Subtropical Horticulture in Lucknow has developed an interspecific hybrid rootstock of guava hybrid Psidium guajava × Psidium molle (PGPM). The rootstock has been demonstrated to be wilt-resistant and grafted successfully with commercial guava varieties [22, 23]. In light of the foregoing, it was hypothesized that the MYB also play a crucial role in determining the peel and pulp colour of guava fruit in PG as well as wilt resistance in PGPM. The first step for alteration via orderly breeding or genome editing is the identification of MYB candidate genes and the controlled pathways [24, 25]. The scarcity of genomic data for guava presents a significant challenge for genetic study. However, the development of genomic and transcriptomic analytical resources and tools is being aided by NGS techniques and bioinformatics pipelines. No attempt has been made to describe MYB genes implicated in guava development, even though physiological studies have been conducted. The objective of this study was to evaluate MYB expression in guava fruit pulp, root, and seed to determine its function by in silico analysis of the root transcriptome data of guava.
Materials and method
Transcriptome sequencing and in silico analysis
On the Illumina platform, paired-end sequencing was used to analyze the transcriptome sequence of the PGPM. The libraries were created using the Illumina TruSeq Stranded Total RNA Library Preparation Kit following the manufacturer's instructions using input total RNA of less than 1 g. Using Trinity software at default settings, high-quality reads were achieved. Trinity software was used with the default parameters to produce high-quality reads. The CD-HIT software was used to further process the transcripts for the prediction of Unigenes. With the help of blast analysis, more than 35,000 CDS were located with precise gene annotations. Nucleotide sequences were analyzed using nucleotide BLAST against mango genome data in the National Center for Biotechnology Information to determine the chromosomal placement and location of MYB CDS (NCBI). Using the online sequence manipulation suite (https://www.bioinformatics.org/sms2/), the nucleotide CDS sequence was converted into amino acid sequences. The MOTIF Search tool (https://www.genome.jp/tools/motif/) was used to analyze the motif search results. Using the MUSCLE tool (https://www.ebi.ac.uk/Tools/msa/muscle/), multiple amino acid sequence alignments of MYB proteins were carried out using CDS of transcriptome data. Weblogo (https://weblogo.berkeley.edu) tool was used to prepare the logo sequences for the R2-R3-MYB domains of MYB proteins. The gene expression analysis of mine MYB family genes was carried out using commonly occurring CDS (based on common NR blast hit accession) taking as control with hybrid Psidium guajava × Psidium molle hybrid (wilt-resistant) as treated. The Gene Ontology (GO) and Subcellular localization analysis of the mine MYB family genes was performed using the online web-server BUSCA (Bologna Unified Subcellular Component Annotator) (https://busca.biocomp.unibo.it/). The phylogenetic position of mine MYB family genes with respect to the reference MYB gene of Arabidopsis thaliana (AAD46772) was performed using MEGA version 5.2 software. The protein sequence of mine MYB family genes was aligned through by inbuilt MUSCLES alignment tool of MEGA version 5.2 software. The evolutionary history was inferred by using the Maximum Likelihood method based on the Poisson correction model with bootstrap replications of 500. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value [26].
Semi-quantitative RT-PCR analysis
Lalit (pulp, seed, and root) and Shweta (pulp) samples of frozen guava were pulverized to a fine powder with liquid nitrogen using a mortar and pestle. Total RNA was isolated using the Spectrum TM plant total RNA kit (Sigma, USA) following the manufacturer's instructions. NanoDrop was used to monitor the purity and quantity of total RNA. The Maxima first strand cDNA synthesis kit for RT-qPCR (Genetix) was used to reverse transcribe 2 g of total RNA from each sample using OligodT and random primers under the manufacturer guidelines. A 1:5 dilution of the synthesized cDNA in nuclease-free water was performed before qRT-PCR analysis. The following parameters were employed to develop primer (Table 1) using IDT PrimerQuest software and the CDS acquired from fruit transcriptome data: OligoAnalyzer was used to check for the presence of stable hairpins and dimers. The ideal length was 25 base pairs, the GC content was 50–55%, the melting point was 57 °C, and the amplicon length range was 100–200 base pairs. For in silico confirmation of each gene’s specificity, the produced primer pair was then aligned to all guava CDS. One hundred nanograms of cDNA, 0.5 µm conc. of each primer, 2.5 mM dNTPs, and 1 unit of Taq DNA polymerase were mixed with 1 × PCR buffer to perform PCR amplification in a total volume of 10 µl. The reaction was run through 35 cycles in a Bio-Rad thermal cycler, starting with an initial denaturation at 94 ℃ for 30 s, followed by 57 °C for 30 s, 72 ℃ for 30 s, and a final extension at 72 °C for 5 min. PCR-amplified products were visualized via a trans-illuminator after being resolved on a 2.0% agarose gel made with 1 × TBE buffer and 0.5 µg of ethidium bromide.
Result and discussion
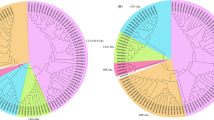
In this study, 2.64 GB of data was generated from root transcriptome analysis of the PGPM. Trinity software was used to do a de novo assembly of high-quality reads, identifying 170,027 transcripts with a maximum length of 962 bp. The top-hit species distribution showed that the species Eucalyptus grandis received the bulk of hits. We have mined 15 different MYB transcription factors genes/transcripts (MYB3, MYB4, MYB5, MYB6, MYB23, MYB44, MYB46, MYB51, MYB86, MYB82, MYB90, MYB114, MYB305, MYB308, and MYB330) in the root transcriptome data of guava. Further, the phylogenetic position of mine MYB family genes with respect to the reference MYB gene of Arabidopsis thaliana (AAD46772) is depicted in Fig. 1. Phylogenetic analysis grouped the mine MYB family genes into four clusters.
Molecular phylogenetic analysis by maximum likelihood method. The tree with the highest log likelihood (-2952.9718) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 65 positions in the final dataset
We used a bioinformatics technique to study domain analysis in coding sequences, which reveals that all mined MYB genes possess the conserved R2-MYB and R3-MYB domains and were found to be unevenly distributed throughout guava chromosomes (Fig. 2; Table 2). WebLogo was used to build sequence logos that could be used to show conservation at specific points. The conserved amino acids shared by MYB domains were strikingly similar, as seen in Figs. 3 and 4. According to these findings, the R2 and R3 repeats of the guava MYBs proteins contain a large number of conserved amino acids, including the distinctive Trp (W). The R2 repeat contains three conserved Trp (W) residues. The R3 repeat only conserved the second and third Trp (W), while the first Trp (W) was frequently replaced with phenylalanine (F) or isoleucine (I) (Figs. 3 and 4). It is possible that substitution at the first Trp (W) residue causes the recognition of new target genes and/or results in a reduction in the DNA-binding activity against target genes. The C-terminal domain is more mutable, whereas the N-terminal domain is more conserved [6, 27]. Gene Ontology (GO) and Subcellular localization analysis of the identified MYB family genes revealed that MYB5, MYB3, MYB1, MYB308, MYB51, MYB86, MYB4, MYB46, MYB23, MYB330, MYB114, MYB305, MYB44 were localized in the nucleus. Whereas the subcellular localization of MYB90 and MYB5 accounted for chloroplast and endomembrane systems respectively (Table 3 and supplementary data).
The expression of MYB3, MYB4, MYB23, MYB86, MYB90, and MYB308 was also evaluated in the Lalit fruit pulp (red pulp), seed, root, and Shweta pulp (white pulp) (Fig. 5). MYB23 and MYB308 were only expressed in the root tissue, indicating that they play a crucial role in the development of roots. While pulp and root tissue showed MYB4 expression (Fig. 4). MYB86 and MYB90 expression was identified in all tissues, including the root, seed, and pulp, indicating that they play a significant role in the development of fruits and roots (Fig. 4). MYB3 was only expressed in the fruit pulp, highlighting its crucial function in fruit ripening (Fig. 4). The heatmap analysis also accounted the exclusive significant higher expression of MYB23 gene in wilt-resistant PGPM as compared with wilt susceptible Psidium guajava (Fig. 6). This result indicates that higher expression of MYB23 (only expressed in root) gene might play a crucial role to confer the wilt resistance in PGPM.
Heatmap of upregulated and downregulated 15 mine MYB gene. Scaling is the base mean value, i.e., dark red colour represents upregulated genes whereas green colour represents downregulated genes. MYB family genes of PG (Psidium guajava) taking as control while PGPM (hybrid Psidium guajava × Psidium molle) is treated
Gene functions can be better understood by understanding the patterns of gene expression. Recent research has revealed that the pear’s skin, bud, and fruit express MYB genes [28, 43]. The MYB gene is responsible for controlling the synthesis of anthocyanins and proanthocyanidins, which affects plant structure and seed colour, respectively [44, 45].
Conclusion
It has been established that members of the MYB gene family play many regulatory roles in controlling how plants respond to diverse biotic and abiotic stresses. Root transcriptome analysis of Psidium guajava × Psidium molle hybrid generated 2.64 GB data. In silico mining including motifs and expression of guava MYBs was carried out in the present study. A total of 15 MYB family members were identified. They were unevenly distributed among chromosomes in guava probably with the occurrence of gene duplication. The exclusive significant higher expression of the MYB23 gene (only expressed in root) in wilt-resistant PGPM indicates that it might play a crucial role to confer the wilt resistance in PGPM. Furthermore, the expression patterns of some MYB demonstrated that MYB might participate in the regulation of fruit ripening, seed and root development. Future guava fruit quality will be enhanced with the help of the identification and study of MYB genes. The insights from these findings may help with future functional studies of the MYB genes to clarify their biological functions in guava.
Availability of data and materials
The data generated during the process of this work are available upon request.
Abbreviations
- PG :
-
Psidium guajava
- PGPM :
-
Hybrid (Psidium guajava × Psidium molle)
- MYB:
-
Myeloblastosis
- DNA:
-
Deoxyribonucleic acid
- CDS :
-
Coding sequence
- NCBI:
-
National Center for Biotechnology Information
- cDNA:
-
Complementary deoxyribonucleic acid
- RT-PCR:
-
Reverse transcription PCR
- Taq:
-
Thermus aquaticus
- TBE:
-
Tris-Borate-EDTA
- Trp:
-
Tryptophan
- WD:
-
Beta-transducin repeat
References
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290(5499):2105–2110. https://doi.org/10.1126/science.290.5499.2105
Ptashne M (1988) How eukaryotic transcriptional activators work. Nature 335:683–689. https://doi.org/10.1038/335683a0
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34. https://doi.org/10.1111/j.1365-313X.2004.02183.x
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. CurrOpin Plant Biol 4:447–456. https://doi.org/10.1016/s1369-5266(00)00199-0
Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He DN, Ogata K, Nishimura Y, Ishii S (1990) The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem 265(32):19990–19995
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581. https://doi.org/10.1016/j.tplants.2010.06.005
Kranz HD, Denekamp M, Greco R, ** H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Weisshaar B (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16(2):263–276. https://doi.org/10.1046/j.1365-313x.1998.00278.x
** H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585. https://doi.org/10.1023/a:1006319732410
Kranz H, Scholz K, Weisshaar B (2000) c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J 21:231–235. https://doi.org/10.1046/j.1365-313x.2000.00666.x
Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A (1996) The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Mol Biol 3:178–87. https://doi.org/10.1038/nsb0296-178
Zhu Y, Dong C, Zhu JK, Hasegawa PM, Bressan RA (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 12:9966–9971. https://doi.org/10.1073/pnas.0503960102
Gao Z, Liu C, Zhang Y, Li Y, Yi K, Zhao X, Cui ML (2013) The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in empire red leaf cotton under light. PLoS ONE 8(10):1–12. https://doi.org/10.1371/journal.pone.0077891
Nimisha S, Kherwar D, Ajay KM, Singh B, Usha K (2013) Molecular breeding to improve guava (Psidiumguajava L.): current status and future prospective. Sci Horti 164:578–588. https://doi.org/10.1016/j.scienta.2013.10.017
Jain SM, Priyadarshan PM (2009) Breeding plantation tree crops: Tropical species. In Annals of botany. New York, USA: Springer 104:vii–xi. https://doi.org/10.1093/aob/mcp206
Pommer CV, Murakami KRN (2009) Breeding Guava (Psidium guajava L.). In: Jain SM, Priyadarshan PM (eds) Breeding Plantation Tree Crops: Tropical Species. Springer, New York. https://doi.org/10.1007/978-0-387-71201-7_3
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970. https://doi.org/10.1093/pcp/pcm066
Rock CD (2013) Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends Plant Sci 18:601–610. https://doi.org/10.1016/j.tplants.2013.07.006
Dhar MK, Sharma R, Koul A, Kaul S (2015) Development of fruit color in Solanaceae: a story of two biosynthetic pathways. Brief Funct Genomics 14:199–212. https://doi.org/10.1093/bfgp/elu018
LeRoy J, Huss B, Creach A, Hawkins S, Neutelings G (2016) Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 7:https://doi.org/10.3389/fpls.2016.00735.
Yao G, Ming M, Allan AC, Gu C, Li L, Wu X (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92:437–451. https://doi.org/10.1111/tpj.13666
Wang N, Xu H, Jiang S, Zhang Z, Lu N, Qiu H (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J 90:276–292. https://doi.org/10.1111/tpj.13487
Rajan S, Kumar R, Mishra AK, Negi SS (2005) Hybrid Guava, Psidium molle × P guajava (INGR No. 04116; IC427822), with Resistance to Guava Wilt. Ind J Plant Genet Resour 18(3):301
Sharma MR, Kumari S, Mishra M (2018) Micropropagation of wilt resistant, inter-specific (Psidium molle × Psidium guajava) rootstock of Guava. Plant Arch 18(1):1170–1174
Mikhaylova EV, Shein MY, Artyukhin AY, Sukhareva AS, Panfilova MA, Kuluev BR (2020) Editing of the MYB genes in Brassica napus as a method to increase anthocyanin pigmentation and stress tolerance. E3S Web Conf 224: 04022.https://doi.org/10.1051/e3sconf/202022404022.
Khusnutdinov E, Sukhareva A, Panfilova M, Mikhaylova E (2021) Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int J Mol Sci 22:8752. https://doi.org/10.3390/ijms22168752
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28:2731–2739
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106. https://doi.org/10.1186/1471-2229-12-106
Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255. https://doi.org/10.1007/s00425-010-1170-5
**e M, Huang Y, Zhang Y, Wang X, Yang H, Yu O (2013) Transcriptome profiling of fruit development and maturation in Chinese white pear (Pyrus bretschneideri Rehd). BMC Genomics 14:823–831. https://doi.org/10.1186/1471-2164-14-823
An JP, An XH, Yao JF, Wang XN, You CX, Wang XF, Hao YJ (2018) BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol 38:1578–1587. https://doi.org/10.1093/treephys/tpy063
Schaart JG, Dubos C, Romero DLI, Van Houwelingen AM, Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria ananassa) fruits. New Phytol 197:454–467. https://doi.org/10.1111/nph.12017
An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169:710–717. https://doi.org/10.1016/j.jplph.2012.01.015
Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140(2):499–511. https://doi.org/10.1104/pp.105.067231
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142(3):1216–1232. https://doi.org/10.1104/pp.106.088104
Li X, Guo C, Ahmad S, Wang Q, Yu J, Liu C, Guo Y (2019) Systematic analysis of myb family genes in potato and their multiple roles in development and stress responses. Biomolecules 9(8):317. https://doi.org/10.3390/biom9080317
Li Z, Peng R, Tian Y, Han H, Xu J, Yao Q (2016) Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol 57(8):1657–1677. https://doi.org/10.1093/pcp/pcw091
He Q, Jones DC, Li W, **e F, Ma J, Sun R, Wang Q, Zhu S, Zhang B (2016) Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci Rep 6:22980. https://doi.org/10.1038/srep22980
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50(4):660–677. https://doi.org/10.1111/j.1365-313X.2007.03078.x
Stracke R, Holtgräwe D, Schneider J, Pucker B, RosleffSörensen T, Weisshaar B (2014) Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biology 14:249. https://doi.org/10.1186/s12870-014-0249-8
Naik J, Rajput R, Pucker B, Stracke R, Pandey A (2021) The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol Biol 106:157–172. https://doi.org/10.1007/s11103-021-01135-x
Wang N, Zheng Y, Duan N, Zhang Z, Ji X, Jiang S (2015) Comparative transcriptomes analysis of redand white-fleshed apples in an F1 population of Malus sieversii f. niedzwetzkyana crossed with M. domestica “Fuji.” PLoS ONE 10:1–20. https://doi.org/10.1371/journal.pone.0133468
** W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14(11):2120–2133. https://doi.org/10.1111/pbi.12568
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in develo** seed. Plant Cell 13:2099–2114. https://doi.org/10.1105/tpc.010098
Wang H, Zhang H, Yang Y, Li M, Zhang Y, Liu J, Dong J, Li J, Butelli E, Xue Z, Wang A, Wang G, Martin C, ** W (2020) The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol J 18(5):1169–1184. https://doi.org/10.1111/pbi.13282
Zheng X, Om K, Stanton KA, Thomas D, Cheng PA, Eggert A, Simmons E, Yuan YW, Conradi Smith GD, Puzey JR, Cooley AM (2021) The regulatory network for petal anthocyanin pigmentation is shaped by the MYB5a/NEGAN transcription factor in Mimulus. Genetics 217(2):36. https://doi.org/10.1093/genetics/iyaa036
Acknowledgements
The author would also like to thank the Director of ICAR-CISH, Lucknow for providing the necessary facilities for the investigation.
Research content
The research content of manuscript is original and has not been published elsewhere.
Funding
The research was funded by the Indian Council of Agricultural Research, New Delhi, India.
Author information
Authors and Affiliations
Contributions
IA and SKS conceived the research, designed experiments. IA and SKS performed the experiments. IA, SKS and MM analyzed the data. IA, SKS, MM and DP prepared the draft. IA and SKS editing and prepared the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have seen the latest version of the manuscript and agree to its publication.
Competing interests
The authors declare that there is no conflict of interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Ancestor chart for GO: 0005634. GO: 0009507. A chlorophyll-containing plastid with thylakoids organized into grana and frets, or stroma thylakoids, and embedded in a stroma. Figure S2. Ancestor chart for GO: 0009507. GO: 0012505. A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or through the use of vesicles. Figure S3. Ancestor chart for GO: 001250. GO: 0005615.That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. Figure S4. Ancestor chart for GO: 0005615.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahmad, I., Soni, S.K., M., M. et al. In-silico mining and characterization of MYB family genes in wilt-resistant hybrid guava (Psidium guajava × Psidium molle). J Genet Eng Biotechnol 21, 74 (2023). https://doi.org/10.1186/s43141-023-00528-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00528-3