Abstract
Background
Dynamic positron emission tomography (PET) images are useful in clinical practice because they can be used to calculate the metabolic parameters (Ki) of tissues using graphical methods (such as Patlak plots). Ki is more stable than the standard uptake value and has a good reference value for clinical diagnosis. However, the long scanning time required for obtaining dynamic PET images, usually an hour, makes this method less useful in some ways. There is a tradeoff between the scan durations and the signal-to-noise ratios (SNRs) of Ki images. The purpose of our study is to obtain approximately the same image as that produced by scanning for one hour in just half an hour, improving the SNRs of images obtained by scanning for 30 min and reducing the necessary 1-h scanning time for acquiring dynamic PET images.
Methods
In this paper, we use U-Net as a feature extractor to obtain feature vectors with a priori knowledge about the image structure of interest and then utilize a parameter generator to obtain five parameters for a two-tissue, three-compartment model and generate a time activity curve (TAC), which will become close to the original 1-h TAC through training. The above-generated dynamic PET image finally obtains the Ki parameter image.
Results
A quantitative analysis showed that the network-generated Ki parameter maps improved the structural similarity index measure and peak SNR by averages of 2.27% and 7.04%, respectively, and decreased the root mean square error (RMSE) by 16.3% compared to those generated with a scan time of 30 min.
Conclusions
The proposed method is feasible, and satisfactory PET quantification accuracy can be achieved using the proposed deep learning method. Further clinical validation is needed before implementing this approach in routine clinical applications.
Similar content being viewed by others
Background
When positron emission tomography (PET) was first proposed, it showed good contrast for performing target area imaging with high quality [1]. As research into fluorodeoxyglucose (FDG) has deepened, the innocuous nature of FDG and its high tumor uptake percentage compared to that of other tissues have allowed PET imaging to show strong tumor diagnosis potential [2]. Chemotherapy and chemoradiotherapy patients are increasingly monitored using PET with 18F-FDG [3]. In routine clinical practice, the standard uptake value (SUV) is highly applied because the glucose metabolic rate and SUV have a good relationship, and this index is easy to obtain [4]. However, the SUVs of static PET images are affected by many different factors, such as the variable uptake period (the time between injection and imaging) and reconstruction parameters (filters, number of iterations, and decay correction) of different scanning instruments, making it problematic to compare SUVs acquired in different places. For this reason, graphical methods such as the Patlak plot are more promising due to their robustness and simplicity in clinical use case [5]. When rigorous and reliable, quantitative analyses can offer more valuable information for clinical practice [6].
Dynamic PET images form a better imaging modality for calculating quantitative values. The first few frames of a dynamic PET image are very short, resulting in considerable noise and a low signal-to-noise ratio (SNR) [7]. Therefore, in most cases, the scanning time required for each time frame of dynamic PET images gradually increases, and the whole process takes at least an hour so that the time activity curve (TAC) is highly accurate. Many parameters can be computed once the TAC is obtained. With the Patlak plot method, the Ki parameter, which is the net uptake rate constant, is used most often. PET Patlak parametric images have been generated based on direct reconstruction using different methods (e.g., the kernel method [8,9,10], deep image prior with the alternating direction of multipliers method (ADMM) [11,12,13,14], the hybrid approach [15], and a method with only a deep network [16]). These methods make the reconstruction process much longer when obtaining parametric images, and some methods do not work well for real patient data due to the fact that they conduct training with simulated data. Therefore, none of these methods can be used in clinical practice. Parametric imaging is time-consuming, and the resulting noisy images require interpretation by skilled users [17]. By reducing the image noise and generation time, parametric images can be made available for clinical use much more quickly. In our study, we made the first attempt to solve this problem. We used only the first 30 min of dynamic PET images. After applying our algorithm, we obtained higher-quality parametric images than those acquired after scanning for 30 min, thus reducing the original 1-h scanning time to half an hour.
Methods
Feature extraction network
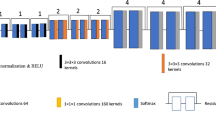
In computer vision, an increasing amount of research points to the importance of convolutional neural networks. Properly trained convolutional neural networks have superior effects in image generation, image segmentation, and other aspects that surpass those of traditional computer vision-based processing methods. At the same time, convolutional neural networks can automatically extract the features of images through training. Based on these studies, we build a fully convolutional neural network with a network architecture that looks like the U-Net architecture that is often used in medical image segmentation. An encoder first downsamples the original 1-channel SUV images. Then, the high-level semantic information of the image is encoded through a series of convolutional or pooling operations to obtain an image feature vector. This feature vector is then sent to a decoder, which returns the information the encoder takes out. This process eliminates noise, which is harder to learn or fit into the network than a useful signal. We change the batch normalization operation in the network to a group normalization operation and add skip connections, similar to those in the residual network, to speed up the training process and improve the quality of the generated images. To be more specific, a basic block called a DoubleConv block makes up many other blocks. The GroupNorm layer and the activation layer come after the two convolutional layers in the DoubleConv block. The rectified linear unit (ReLU) function is chosen as the activation function. The number of channels per group is set to 16 for GroupNorm. The encoder comprises one DoubleConv block and four DownConv blocks that are made up of one maximum pooling layer and one DoubleConv block. The Maxpool layer’s function is to perform downsampling by a factor of 2. The first DoubleConv block maps the channel size of the input image to the target channel size for the subsequent calculations. The target channel sizes of the blocks are set to 64, 128, 256, 512, and 1024, which means that the output feature image size is 1/16 of the original image size. The decoder then takes the last feature image to perform upsampling 4 times using UpConv blocks. Each UpConv block comprises one transposed convolutional layer and one DoubleConv block. For each block at the same level, skip connections are made between the encoder and decoder. The architecture of the feature extraction network is shown in Fig. 1. The dimensionality of the input is described in the “Training Setup” section, and the flow of data from one network to the other is shown in Fig. 2. In Fig. 2, we refer to the following kinetic model network as a pointwise neural network (Fig. 3).
The relation between the feature extraction network and the kinetic model network (pointwise neural network). We obtained each frame’s feature maps using U-Net. All 220 feature maps were then input into a pointwise neural network, which was implemented by a stack of convolutions with a kernel size of 1. Therefore, each voxel was a 220-dim vector that was fed into the kinetic model network
Kinetic model network
The physiological system of dynamic processes in the tissue of interest is decomposed into several compartments, which interact with each other. In PET, tracer kinetic modeling is based on compartmental analysis. Ordinary differential equations (ODEs) continuously and deterministically represent the compartmental system. Each equation describes the temporal rate of change exhibited by the material in a compartment. These rates of change are controlled by the physical and chemical rules that govern how materials move from one compartment to another. These rules include diffusion, temperature, and chemical reactions [18]. The vast majority of articles use the 2-tissue compartment model (2TCM) with the Patlak method to analyze dynamic PET images. Since most researchers have looked into the 2TCM and found that it works [7], our method also builds the network on the 2TCM. The ODEs of the 2TCM are described as follows:
where \({K}_{1}\) is a constant that represents the rate of influx from plasma to tissue, \({k}_{2}\) is a constant that represents the rate of efflux from the first compartment in the 2TCM, \({k}_{3}\) is the rate of transfer from a nonspecific compartment to a specific compartment in a reversible or irreversible 2TCM, and \({k}_{4}\) is the rate of transfer from a specific compartment to a nonspecific compartment in the reversible 2TCM. To increase the complexity and diversity of the TACs generated by the network, we do not fix \({k}_{4}\) as 0. However, the network is capable of generating TACs when \({\mathrm{k}}_{4}\) is equal to 0. \({C}_{0}(t)\) is the input blood function, \({C}_{1}\left(t\right)\) is the concentration of the nondisplaceable compartment, and \({C}_{2}\left(t\right)\) is the concentration of the binding radiotracer in the specific compartment; the tissue concentration \({C}_{\mathrm{T}}(t)\) is the sum of the nondisplaceable and specific compartment concentrations. [19].
The solution of these ODEs is the convolution of an exponential function with the input function. The equations are as follows:
The total activity concentration (e.g., in nCi/ml) for a voxel at a given time is denoted by
where \({\mathrm{\varphi }}_{\mathrm{s}}\) represents the parameters of the kinetic model. The volume fraction of a voxel that is made up of blood is denoted by the constant \({f}_{v}\). \({C}_{\mathrm{WB}}\)(nCi/ml) is the concentration of tracer activity in whole blood (i.e., plasma plus blood cells plus other particulate matter) [20].
Our method uses the blood input function \({C}_{0}\left(t\right)\) as the whole blood function \({C}_{\mathrm{WB}}(t)\). We form the kinetic model network, a convolutional neural network with 1 × 1 convolutional layers that let each voxel be computed separately while reducing the number of parameters and increasing the training speed of the network. We applied the feature extraction network to each individual time frame image of the dynamic PET data, extracting 10 feature maps for each time frame. With a total of 22 time frames, there are 220 feature maps in total. This means that each voxel is represented by a 220-dimensional vector, as shown in Fig. 2. The feature extraction network's output feature vectors are fed into the kinetic model network. Moreover, the kinetic model network predicts the five parameters (\({f}_{v},{K}_{1},{k}_{2},{k}_{3},{k}_{4})\) of the 2TCM for each voxel based on the inputs. After obtaining the parameters generated by the network, we can use these parameters to obtain the whole TAC through the 2TCM. Once we have obtained the time activity curves for each voxel, we can calculate the dynamic PET image at any desired time frame. To be more specific, the intensity of the image at pixel j in time frame m, \({x}_{m}({\theta }_{j})\), is determined by
where \({t}_{m,s}\) represents the starting time of frame m, \({t}_{m,e}\) represents the ending time of frame m, and \(\lambda\) represents the decay constant of the radiotracer. \({C}_{\mathrm{PET}}\left(t;{\theta }_{j}\right)\) denotes the tracer concentration in pixel j at time t, which is determined using the aforementioned kinetic model with the parameter vector \({\theta }_{j}\in {R}^{{n}_{k}\times 1}\).
Then, we can estimate Ki using the Patlak plot method:
where \({K}_{i}\) is the constant rate of irreversible binding. \({V}_{0}\) is the distribution volume of the nonspecifically bounded tracer in the tissue. \(x(t)\) is the integrated activity of the tissue up to time t. \({C}_{p}(t)\) is the plasma concentration of the tracer at time t.
Training setup
Only the dynamic images obtained during the first thirty minutes were fed into the whole neural network. All the inputs were normalized to SUV images. The input matrix had a shape of \({T}_{i}\) ×1 × H × W, where \({T}_{i}\) corresponds to the total number of time frames within the initial thirty minutes. H and W denote the height and width of the image, respectively. The number of input channels was specified as 1. The size of the output matrix, representing the whole network's output, was \(T\times H\times W\), where T represents the number of time frames in the dynamic PET images. Furthermore, the loss function was the Huber loss [21], which is very resistant to outliers.
To train the kinetic model network, we calculated the loss between the generated images and the ground truth as the loss function.
where \({\mathrm{pred}}_{\mathrm{SUV}}^{t}(x,y)\) is the pixel value of the generated image at position (x,y) for the t-th frame. \({\mathrm{gt}}_{\mathrm{SUV}}^{t}(x,y)\) is the pixel value of the ground truth at position (x,y) for the t-th frame. T is the total number of frames. M and N are the height and width of the images, respectively. Due to the fact that the first 30 min of dynamic PET images (the first 22 frames) were already used as network inputs, we only utilized the images from the subsequent 30 min (last 6 frames) as the training targets.
Additionally, we added a time difference loss function for the linear part of the Patlak model.
where \({C}_{p}(t)\) is the blood input function. \(K(x,y)\) is the Ki parameter of the Patlak plot at position (x,y). x(t), y(t), and \({\mathrm{diff}}_{k}(\cdot )\) are defined according to the definitions provided in Eq. (8). \({T}_{\mathrm{linear}}\) is the total number of frames that represent the linear portion in the Patlak plot model. \({t}_{k}\) represents the kth time frame.
Thus, the total loss is as follows:
where \(\lambda\) is a hyperparameter that adjusts the weight between fitting Ki and the SUV.
We did not train the feature extraction network and the kinetic model network separately; instead, we treated them as one end-to-end network and trained them together. The optimizer was chosen as adaptive moment estimation (Adam) [22], and the learning rate was set to 1e-4. We used a strategy that adjusted the learning rate to one-tenth of the original value every 10,000 iterations, with a lower bound of 1e-7. We trained our network on an NVIDIA GeForce RTX 3090 GPU for a total of 10 epochs. Each epoch included 7171 iterations. To validate the effectiveness of our proposed method that incorporates a kinetic model, we compared it to a method with the exact same network structure but without the kinetic model. In other words, we directly predicted the SUV images for the last 30-min time frames without the need to perform the steps of the kinetic model. Additionally, while maintaining the rest of the network architecture unchanged, we removed the sigmoid activation function from the final layer. We are still employing a point-wise neural network approach. We refer to this method as "without model" in the figure. The method without incorporating the kinetic model adopted the same hyperparameter settings and loss function as the full model. This was done to minimize the influence of other factors and ensure the accuracy of the conclusions.
Patient PET data
The network's training dataset was obtained from the Cancer Hospital of the Chinese Academy of Medical Sciences Shenzhen Center, which included 7313 slices of data from 103 patients acquired with the GE Healthcare Discovery MI Dr PET/CT Scanner. All patients had space-occupying lung lesions, which can also be called pulmonary nodules. Both benign and malignant lesions were present. We randomly selected 10 patients as the test set and 93 patients as the training set. The patient's height range was 1.641 m ± 0.089 m, and the weight range was 63.0 kg ± 10.36 kg. Information on the patient's gender and age were unavailable because the patient's data were anonymized and desensitized. The dynamic PET data were divided into 28 frames: 6 × 10 s, 4 × 30 s, 4 × 60 s, 4 × 120 s, and 10 × 300 s with total radionuclide doses of \({\mathrm{F}}^{18}\)-FDG ranging from 201.83 Mbq to 406.46 Mbq for different patients. Each time frame of the dynamic PET data was an image array of 256 × 256 × 71 voxels with a voxel size of 1.95 × 1.95 × 2.79 mm3. The blood input function was manually extracted from the image region of the descending aorta.
Results
Qualitative image quality assessment
Figure 4 shows that the overall visual effect of the generated images was close to that of the reference images and presented most of the anatomical structure details, which was based on the observations of the three views in the coronal, sagittal, and transverse planes. In addition, some high-uptake regions could still be effectively represented in the generated images. A better SNR could be obtained using our proposed method, which also had a positive effect in terms of noise reduction for improving the image quality. Figure 5 shows that a noisy Ki image would have been obtained if we applied the Patlak plot method on the first 30 min of dynamic PET data. However, we can see that the noise level was reduced through our method, and we could generate a more reasonable Ki image. Our method could show more anatomical details of tissues and organs than the no-kinetic-model network. Both our network and the no-kinetic-model network exhibit artifacts in the cardiac region in Figs. 4 and 5. This phenomenon is likely attributed to the fact that the network's input consists of various time frames from the initial 30 min. Due to cardiac motion between time frames, the lack of consistency in features extracted by the feature extraction network introduces significant noise, resulting in the appearance of artifacts. Figure 6 shows that our method gave more accurate SUV results in most tissue regions, but it provided SUVs that were lower than the real values in some metabolically active areas that did not fit the kinetic model. However, if we did not have kinetic models, our networks may have produced very inaccurate predictions about some tissues and organs. This would make the images less useful for diagnosis. Concerning the Ki image, the original Ki image generated in the first 30 min was predicted accurately for the hypermetabolic region because the TAC of the hypermetabolic region showed an upward trend in the early stage and quickly entered the linear stage on the Patlak plot. The Ki image acquired by our method presents the same conclusions as the SUV forecast.
Examples of generated dynamic PET images obtained from different planes (the transverse plane, coronal plane and sagittal plane). The ground truths are the last frames of the dynamic PET images, which were obtained 1 h after injection with a 5-min scanning time. The images in the upper-right corner were obtained by our proposed network with the kinetic model, and the images on the bottom were generated by the same network without the kinetic model
Example of generated parametric Ki images obtained from different planes (the transverse plane, coronal plane and sagittal plane). The Ki images of both the ground truth and our proposed method were obtained by fitting the last 13 frames of the Patlak plot’s data points with linear regression. The main differences between both methods are that the data points from the last 30 min were generated by the proposed network rather than being real. The Ki images of the method without the kinetic model in the lower right were generated directly rather than by fitting a Patlak plot. The images in the upper right were obtained by fitting only the first 30 min of frames
Quantitative image quality assessment
We compared the image evaluation metrics computed by various deep learning methods, such as the attention-based hybrid image quality (AHIQ) method [23], the deep image structure and texture similarity (DISTS) approach [24], and the learned perceptual image patch similarity (LPIPS) technique [25], and some metrics without deep learning, such as gradient magnitude similarity deviation (GMSD) [26], most apparent distortion (MAD) [27], the normalized Laplacian pyramid distance (NLPD) [28], and the visual saliency-induced index (VSI) [29]. We also included traditional metrics such as the structural similarity index measure (SSIM), the peak SNR (PSNR), the normalized mutual information (NMI), and their improved versions such as the multiscale SSIM (MS-SSIM) [30], information content-weighted SSIM (IW-SSIM) [31], feature similarity index measure (FSIM) [32], spectral residual-based similarity index measure (SR-SIM) [33], discrete cosine transform (DCT) subband similarity (DSS) [34], and Haar perceptual similarity index (HaarPSI) [35] (Figs. 7, 8). These measurement methods showed that, on average, our method worked better than the Ki images made from 30 min of dynamic PET images.
A comparison of the image quality of the SUV images produced with and without a kinetic model, where a real 1-h static PET image was used as a reference. The suffix “_h” means that the higher this metric is, the better the image quality. In contrast, the suffix “_l” means that this metric is a distortion index, so the lower this metric is, the better the image quality. The assessment of the radar image on the right shows that the larger the footprint is, the better the image quality
Image quality assessment of the Ki images generated by different methods with the same input. The explanations and descriptions of the pictures are the same as those in Fig. 7
Figure 9 shows that our method produced consistently better NMI metrics for all 10 patients' data when the ground truth was used as the reference image. This goes some way toward explaining the usability of our approach. Figure 10 shows that, except for the eighth patient, our method yielded better PSNR measurements than the original method. Figure 11 shows that the SSIM decreased significantly if the parameter image was made directly without using a kinetic model. However, this problem did not occur with our proposed method, and it can be seen that our method obtained better SSIMs for all patients except for patient 8.
To determine how close the synthetic Ki images were to the real images, a test subject with a malignant lung tumor was chosen from the test data. The region of interest (10 × 10 × 10) of the subject's tumor was delineated and analyzed in a Bland‒Altman plot. The Bland‒Altman plots showed that the 95% limits of agreement between the ground truths and the Ki images synthesized by the algorithm in this paper were between -0.029 ~ 0.03 (mean: 0.00), and the 95% limits of agreement between the ground truths and this Ki images synthesized by the method without incorporating the kinetic model were between − 0.027 and 0.034 (mean: 0.003), which were slightly larger than those of our proposed network. The 95% limits of agreement between the Ki images generated only with the original data acquired in the first 30 min and the ground truths were between − 0.029 and 0.039 (mean: 0.005), presenting the largest error.
Discussion
We developed a new way to quickly and effectively combine deep learning with kinetic models to form dynamic PET images for the next 30 min from the dynamic PET images of the first 30 min. This method is the first time that SUV and Ki parametric images have been made at the same time, and it works well. Real patient data were used to show that the proposed method can make parametric images that match the reference images derived from Patlak plots. By using different metrics, such as evaluation criteria involving deep learning and metrics using the traditional computational method of extracting texture features for evaluating image quality, we showed that the image quality generated by our deep learning method combined with a kinetic model is better for Ki parameter images. This development may significantly reduce the required scanning time and improve patient comfort.
According to our observations, the SUV images generated by our method contained a certain amount of dynamic PET trend information for the first 30 min while bound by the curve of the kinetic model. If the target tissue's TAC does not fit the current kinetic model, it will not be suitable for constructing highly accurate parametric and SUV images. Additionally, because the generated images are learned from the input of the dynamic PET SUV source, if the input source does not contain the trend of the next 30 min, then the images will not be generated well either.
The Ki images generated by directly using a deep learning approach cannot guarantee consistency with the real situation, which can be seen in the SSIM metric comparison (Fig. 10), and the interpretability of deep learning is very low, which limits the application of deep learning in the medical field. Our method uses a kinetic model to make deep learning more interpretable to a certain degree.
The deep learning framework we proposed is also scalable. In future, as the level of pharmacokinetic modeling of human tissues and our understanding of how human tissues work metabolically improve, the TACs made with our method will become more accurate.
Conclusion
In this work, we looked at an approach that combines kinetic models with deep learning using only the first 30 min of dynamic PET images to obtain the next 30 min of dynamic PET images and parametric Ki images. On data acquired from 103 patients, deep learning techniques combined with kinetic models were evaluated in terms of subjective and objective measures. The results showed that accurate parametric Ki image estimation is valid, can reduce the required scanning time and can make patients more comfortable. Although the proposed method performed well in quantitative evaluations, further validation is needed in clinical applications. In future, more research should be done on the kinetic modeling process to improve the performance of the existing models. For example, pharmacokinetic models that work for both tumors and normal tissues could be studied to make neural network models much more accurate.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- PET:
-
Positron emission tomography
- SUV:
-
Standard uptake value
- SNR:
-
Signal-to-noise ratio
- TAC:
-
Time activity curve
- SSIM:
-
Structural similarity index measure
- PSNR:
-
Peak signal-to-noise ratio
- RMSE:
-
Root mean square error
- FDG:
-
Fluorodeoxyglucose
- ADMM:
-
Alternating direction of multipliers method
- ReLU:
-
Rectified linear unit
- ODE:
-
Ordinary differential equation
- 2TCM:
-
2-Tissue compartment model
- Adam:
-
Adaptive moment estimation
- AHIQ:
-
Attention-based hybrid image quality
- DISTS:
-
Deep image structure and texture similarity
- LPIPS:
-
Learned perceptual image patch similarity
- GMSD:
-
Gradient magnitude similarity deviation
- VSI:
-
Visual saliency-induced index
- MAD:
-
Most apparent distortion index
- NLPD:
-
Normalized Laplacian pyramid distance
- NMI:
-
Normalized mutual information
- MS-SSIM:
-
Multiscale SSIM
- IW-SSIM:
-
Information content-weighted SSIM
- FSIM:
-
Feature similarity index measure
- SR-SIM:
-
Spectral residual-based similarity index measure
- DCT:
-
Discrete cosine transform
- DSS:
-
DCT subband similarity
- HaarPSI:
-
Haar perceptual similarity index
References
Jones T, Townsend DW. History and future technical innovation in positron emission tomography. J Med Imaging. 2017;4: 011013.
Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021;71:333–58.
Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol. 2015;I(42):328–54.
Farwell MD, Pryma DA, Mankoff DA. PET/CT imaging in cancer: current applications and future directions. Cancer. 2014;120:3433–45.
Tomasi G, Turkheimer F, Aboagye E. Importance of quantification for the analysis of PET data in oncology: review of current methods and trends for the future. Mol Imaging Biol. 2012;14:131–46. https://doi.org/10.1007/s11307-011-0514-2.
Galli G, Indovina L, Calcagni ML, Mansi L, Giordano A. The quantification with FDG as seen by a physician. Nucl Med Biol. 2013;40:720–30. https://doi.org/10.1016/j.nucmedbio.2013.06.009.
Dimitrakopoulou-Strauss A, Pan LY, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol. 2021;I(48):21–39. https://doi.org/10.1007/s00259-020-04843-6.
Gong K, Wang GB, Chen KT, Catana C, Qi JY. Nonlinear PET parametric image reconstruction with MRI information using kernel method. Proc Spie. 2017. https://doi.org/10.1117/12.2254273.
Gong K, Cheng-Liao JX, Wang GB, Chen KT, Catana C, Qi JY. Direct Patlak reconstruction from dynamic PET data using the kernel method with MRI information based on structural similarity. IEEE Trans Med Imaging. 2018;37:955–65. https://doi.org/10.1109/Tmi.2017.2776324.
Mao X, Zhao S, Gao D, Hu Z, Zhang N. Direct and indirect parameter imaging methods for dynamic PET. Biomed Phys Eng Express. 2021. https://doi.org/10.1088/2057-1976/ac086c.
Gong K, Catana C, Qi JY, Li QZ. Direct reconstruction of linear parametric images from dynamic PET using nonlocal deep image prior. IEEE T Med Imaging. 2022;41:680–9. https://doi.org/10.1109/Tmi.2021.3120913.
Gong K, Catana C, Qi JY, Li QZ. Direct patlak reconstruction from dynamic PET using unsupervised deep learning. In: 15th international meeting on fully three-dimensional image reconstruction in radiology and nuclear medicine. 2019;11072. Artn 110720r. https://doi.org/10.1117/12.2534902.
Cui JN, Gong K, Guo N, Kim K, Liu HF, Li QZ. Unsupervised PET logan parametric image estimation using conditional deep image prior. Med Image Anal. 2022. https://doi.org/10.1016/j.media.2022.102519.
Cui JA, Gong K, Guo N, Kim K, Liu HF, Li QZ. CT-guided PET parametric image reconstruction using deep neural network without prior training data. In: Medical imaging 2019: physics of medical imaging. 2019;10948. Artn 109480z. https://doi.org/10.1117/12.2513077.
**e NB, Gong K, Guo N, Qin ZX, Wu ZF, Liu HF, et al. Rapid high-quality PET Patlak parametric image generation based on direct reconstruction and temporal nonlocal neural network. Neuroimage. 2021. https://doi.org/10.1016/j.neuroimage.2021.118380.
Li Y, Hu J, Sari H, Xue S, Ma R, Kandarpa S, et al. A deep neural network for parametric image reconstruction on a large axial field-of-view PET. Eur J Nucl Med Mol I. 2022. https://doi.org/10.1007/s00259-022-06003-4.
Dimitrakopoulou-Strauss A, Pan LY, Sachpekidis C. Parametric imaging with dynamic PET for oncological applications: protocols, interpretation, current applications and limitations for clinical use. Semin Nucl Med. 2022;52:312–29. https://doi.org/10.1053/j.semnuclmed.2021.10.002.
Wang Y, Li E, Cherry SR, Wang G. Total-body PET kinetic modeling and potential opportunities using deep learning. PET Clin. 2021;16:613–25.
Gallezot JD, Lu Y, Naganawa M, Carson RE. Parametric imaging With PET and SPECT. IEEE Trans Radiat Plasma Med Sci. 2020;4:1–23. https://doi.org/10.1109/TRPMS.2019.2908633.
Yokota T, Kawai K, Sakata M, Kimura Y, Hontani H. Dynamic PET image reconstruction using nonnegative matrix factorization incorporated with deep image prior. In: Proceedings of the IEEE/CVF international conference on computer vision; 2019. p. 3126–35.
Oksuz K, Cam BC, Kalkan S, Akbas E. Imbalance problems in object detection: a review. IEEE Trans Pattern Anal Mach Intell. 2020;43:3388–415.
Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014. p. https://arxiv.org/abs/1412.6980
Lao SS, Gong Y, Shi SW, Yang SD, Wu TH, Wang JH, et al. Attentions Help CNNs See Better: Attention-based Hybrid Image Quality Assessment Network. In: 2022 Ieee/Cvf conference on computer vision and pattern recognition workshops (Cvprw 2022). 2022:1139–48. https://doi.org/10.1109/Cvprw56347.2022.00123.
Ding K, Ma K, Wang S, Simoncelli EP. Image quality assessment: Unifying structure and texture similarity. IEEE Trans Pattern Anal Mach Intel. 2020;44(5):2567–81.
Zhang R, Isola P, Efros AA, Shechtman E, Wang O. The unreasonable effectiveness of deep features as a perceptual metric. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. p. 586–95.
Xue W, Zhang L, Mou X, Bovik AC. Gradient magnitude similarity deviation: a highly efficient perceptual image quality index. IEEE Trans Image Process. 2013;23:684–95.
Larson EC, Chandler DM. Most apparent distortion: full-reference image quality assessment and the role of strategy. J Electron Imaging. 2010;19: 011006.
Laparra V, Ballé J, Berardino A, Simoncelli EP. Perceptual image quality assessment using a normalized Laplacian pyramid. Electron Imaging. 2016;2016:1–6.
Zhang L, Shen Y, Li H. VSI: A visual saliency-induced index for perceptual image quality assessment. IEEE Trans Image Process. 2014;23:4270–81.
Wang Z, Simoncelli EP, Bovik AC. Multiscale structural similarity for image quality assessment. In: The thrity-seventh asilomar conference on signals, systems and computers, 2003; 2003. Vol. 2. p. 1398–402.
Wang Z, Li Q. Information content weighting for perceptual image quality assessment. IEEE Trans Image Process. 2011;20:1185–98. https://doi.org/10.1109/TIP.2010.2092435.
Zhang L, Zhang L, Mou X, Zhang D. FSIM: a feature similarity index for image quality assessment. IEEE Trans Image Process. 2011;20:2378–86. https://doi.org/10.1109/TIP.2011.2109730.
Zhang L, Li H. SR-SIM: a fast and high performance IQA index based on spectral residual. In: 2012 19th IEEE international conference on image processing; 2012. p. 1473–6.
Balanov A, Schwartz A, Moshe Y, Peleg N. Image quality assessment based on DCT subband similarity. In: 2015 IEEE international conference on image processing (ICIP); 2015. p. 2105–9.
Reisenhofer R, Bosse S, Kutyniok G, Wiegand T. A Haar wavelet-based perceptual similarity index for image quality assessment. Signal Process Image Commun. 2018;61:33–43.
Acknowledgements
We would like to express our deepest gratitude to the doctors who have generously provided us with the data used in this research. Their invaluable contributions made this study possible. We would also like to thank our supervisor for their guidance and support throughout the research process. Without their expertise and mentorship, this project would not have been possible.
Funding
This work was supported by the National Natural Science Foundation of China (32022042) and the Shenzhen Excellent Technological Innovation Talent Training Project of China (RCJC20200714114436080).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and design of the study. Material preparation and data collection were performed by XW, YZ and JZ. Data analysis and modeling were performed by GL. The first draft of the manuscript was written by GL, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liang, G., Zhou, J., Chen, Z. et al. Combining deep learning with a kinetic model to predict dynamic PET images and generate parametric images. EJNMMI Phys 10, 67 (2023). https://doi.org/10.1186/s40658-023-00579-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40658-023-00579-y