Abstract
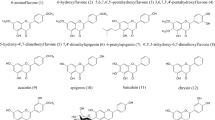
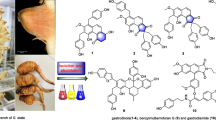
Vibrio species (Vibrio sp.) is a class of Gram-negative aquatic bacteria that causes vibriosis in aquaculture, which have resulted in big economic losses. Utilization of antibiotics against vibriosis has brought concerns on antibiotic resistance, and it is essential to explore potential antibiotic alternatives. In this study, seven compounds (compounds 1–7) were isolated from the Arctic endophytic fungus Penicillium sp. Z2230, among which compounds 3, 4, and 5 showed anti-Vibrio activity. The structures of the seven compounds were comprehensively elucidated, and the antibacterial mechanism of compounds 3, 4, and 5 was explored by molecular docking. The results suggested that the anti-Vibrio activity could come from inhibition of the bacterial peptide deformylase (PDF). This study discovered three Penicillium-derived compounds to be potential lead molecules for develo** novel anti-Vibrio agents, and identified PDF as a promising antibacterial target. It also expanded the bioactive diversity of polar endophytic fungi by showing an example in which the secondary metabolites of a polar microbe were a good source of natural medicine.
Graphical Abstract

Similar content being viewed by others
Introduction
Vibrio species (Vibrio sp.) are a class of Gram-negative aquatic bacteria characterized by motile rods and facultative anaerobic metabolism. They reside in estuarine and coastal environments as well as freshwaters and cause vibriosis that leads to big yield losses in aquaculture (Baker-Austin et al. 2018; Brumfield et al. 2021; Ghosh et al. 2021). V. parahaemolyticus and V. alginolyticus are the most common infectious bacteria in aquaculture (Abd El Tawab et al. 2018), while V. cholerae, V. vulnificus and V. parahaemolyticus are also serious foodborne pathogens that cause gastroenteritis, wound infections, and septicaemia in humans (Ina-Salwany et al. 2019).
In both clinical medicine and aquaculture, antibiotics like quinolone, tetracycline, and erythromycin have been used to control vibriosis (Handayani et al. 2022). With extensive usage of antibiotics, the issue of antibiotic resistance has emerged with complicated cases (Hussain et al. 2020; Li et al. Optical rotations were measured using an Autopol I polarimeter (Rudolph Research Analytical Inc, Boston, MA, USA) in methanol (MeOH). UV spectra were recorded on a Shimadzu UV-1800 spectrophotometer (Shimadzu Corporation Co Ltd, Tokyo, Japan) in MeOH. 1D NMR spectra were recorded on a Bruker A VIII-600 NMR spectrometer using tetramethylsilane as an internal standard. Column chromatography was performed on silica gel (200–300 mesh, Qingdao Marine Chemical Inc, Qingdao, China) and ODS (50 µm, YMC Co Ltd, Kyoto, Japan) on a Flash Chromatograph System (SepaBen machine, Santai Technologies Inc, Changzhou, China). Preparative high-performance liquid chromatography (Pre-HPLC) was performed on a Shimadzu LC-20 system (Shimadzu Co Ltd, Tokyo, Japan) equipped with a Shim-pack RP-C18 column (20 × 250 mm, 10 µm, Shimadzu Co Ltd, Tokyo, Japan) with a flow rate of 10 mL/min at 25 °C. Endophytic fungi species were isolated from the soil samples collected from Arctic Svalbard Archipelago (73–80 °N, 18–31 °E). The fungal isolates were grown in a 15-mL potato dextrose broth for 3 days at 28 °C, and the mycelia were filtered and used for DNA extraction. Fungal isolates were identified by sequencing the internal transcribed regions (ITS). Primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) were used for sequencing the ITS of the fungal genome (Shanghai Personal Biotechnology Co Ltd, Shanghai, China). The sequencing results were aligned with fungal ITS sequences on NCBI using BLASTN. The fungus was identified to be a Penicillium sp. (GenBank no. OP536848) according to the sequence similarity (Schoch et al. 2012). The fungus was incubated on a potato dextrose agar (PDA) medium at 28 °C for 3 days, and the fungal mycelia were inoculated into a 250-mL Erlenmeyer flask containing 100 mL of the potato dextrose broth and fermented at 28 °C with 220 rpm. After 2 days of fermentation, the seed culture (~ 15 mL) was inoculated into an Erlenmeyer flask which contained 100 g of sterilized dry rice and 110 mL of distilled water. All the flasks were stacked at room temperature for 14 days. The fermented rice medium was extracted for three times using ethyl acetate (EtOAc), and the solvents were concentrated by a rotary evaporator to get a crude extract (43.2 g). The crude extract was loaded onto a silica gel column and eluted sequentially with a series of polar solvents, petroleum ether, dichloromethane (CH2Cl2), EtOAc, and MeOH. The EtOAc fraction was partitioned by an ODS column eluted with a gradient of MeOH–H2O (30–100% MeOH) and divided into five fractions, A to E. Based on TLC analysis, fractions B and E were chosen for further purification. Fraction B was purified by an ODS column (acetonitrile–H2O, 35:65) and a semi-preparative HPLC with 60% MeOH/H2O isocratic elution. Compounds 1 (8.2 mg), 2 (10.1 mg), 4 (18.6 mg), 6 (17.9 mg), and 7 (6.3 mg) were obtained from fraction B. Fraction E was purified by an ODS column (acetonitrile–H2O, 30:70) and a semi-preparative HPLC with 55% MeOH/H2O. Compounds 3 (6.1 mg) and 5 (14.5 mg) were obtained from fraction E. The seven compounds (1–7) were dissolved in MeOH and tested with high-resolution electrospray ionization mass spectroscopy (HRESIMS) to obtain the molecular formulas. To determine the 2D structures, compounds were dissolved in deuterated chloroform (CDCl3) (compound 1), deuterated methanol (MeOD) (compounds 2, 3, 4, and 5), or deuterated dimethyl sulfoxide (DMSO-d6) (compounds 6 and 7) for NMR spectrometry. Compounds 2, 4, 5 and 6 were dissolved in MeOH and tested on an Autopol I polarimeter for the optical rotations. V. parahaemolyticus, V. cholerae, V. vulnificus and V. alginolyticus were cultured on 3% NaCl-LB medium (5 g/L yeast extract, 10 g/L peptone, 30 g/L NaCl, pH 7.4) plates at 30 °C for 12 h. The single colony of each strain was inoculated into a 250-mL Erlenmeyer flask containing 100 mL of the NaCl-LB liquid medium and cultured at 37 °C overnight with 200 rpm shaking. The bacterial cultures were diluted to 106 CFU/mL with the NaCl-LB liquid medium for testing. Streptomycin and the seven compounds were dissolved in dimethyl sulfoxide (DMSO) to make 5-mM stocks. The stocks were diluted to desired concentrations with the NaCl-LB liquid medium for testing. Into each well of a 96-well plate, 100 μL of the 106 CFU/mL bacterial dilution and 100 μL of the compound solution with the desired concentration was added. Concentrations of the compounds tested were 500, 250, 125, and 62.5 μM. The test was conducted at 30 °C overnight with three independent repeats. Three-dimensional (3D) crystal structures or homology models of the Vibrio-specific targets reported in recent years on RCSB Protein Data Bank (PDB) or UniProt Database were used for molecular docking (Bonardi et al. 2021; Ding et al. 1993, 1994). In this study, the firm binding of the active compounds to PDF may have changed the protein conformation and affected the activity. After PDF inactivation, the formyl group of the nascent peptide chain in Vibrio cells could not be removed smoothly. This indicates the compounds can inhibit bacterial growth by preventing bacterial protein synthesis (Durand et al. 1999). Moreover, the result showed that electron-withdrawing epoxy units are more likely to form hydrogen bonds than π-rich double bond when binding to the ligand pockets. The discovery that PDF is present in several human parasites also suggests it to be a potential target for antiparasitic agents (Meinnel 2000). In conclusion, seven compounds were isolated from the Arctic endophytic fungus Penicillium sp. Z2230. Three compounds, viridicatol (3), cyclopenol (4) and cyclopenoin (5), showed potent anti-Vibrio activity at micromolar level. Molecular docking of the compounds suggested that the anti-Vibrio activity could come from the inhibition of bacterial peptide deformylase (PDF). These Penicillium-derived compounds are potential lead molecules for the development of novel anti-Vibrio agents. The data also indicate that PDF is a promising target for new antibacterial agents. This study expands the biologically active diversity of polar endophytic fungi, and shows an example in that the secondary metabolites of polar microbes are a good source of natural medicine.Materials and methods
General experimental procedures
Identification of the fungus
Fermentation, extraction, and isolation of fungal compounds
Structure determination of the compounds
Anti-Vibrio activity
Molecular docking
Availability of data and materials
All data generated or analyzed during this study are included in this research article.
References
Abdeltawab AA, Ibrahim AM, Sittien AS (2018) Phenotypic and genotypic characterization of Vibrio species isolated from marine fishes. Benha Veterinary Medical Journal 34(1):79–93
Bai J, Zhang P, Bao GH, Gu JG, Han LD, Zhang LW, Xu YQ (2018) Imaging mass spectrometry-guided fast identification of antifungal secondary metabolites from Penicillium polonicum. Appl Microbiol Biotechnol 102(19):8493–8500
Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J (2018) Vibrio spp. infections. Nat Rev Dis Primers 4(1):1–19
Bonardi A, Nocentini A, Osman SM, Alasmary FA, Almutairi TM, Abdullahc DS, Gratteri P, Supuran CT (2020) Inhibition of α-, β- and γ-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae with aromatic sulphonamides and clinically licenced drugs—a joint docking/molecular dynamics study. J Enzyme Inhib Med Chem 36(1):469–479
Brumfield KD, Usmani M, Chen KM, Gangwar M, Jutla AS, Huq A, Colwell RR (2021) Environmental parameters associated with incidence and transmission of pathogenic Vibrio spp. Environ Microbiol 23(12):7314–7340
Brunati M, Rojas JL, Sponga F, Ciciliato I, Losi GE, Hoog S, Genilloud O, Marinelli F (2009) Diversity and pharmaceutical screening of fungi from benthic mats of Antarctic lakes. Mar Genomics 2(1):43–50
Ding LJ, **ao SJ, Liu D, Pang WC (2018) Effect of dihydromyricetin on proline metabolism of Vibrio parahaemolyticus: inhibitory mechanism and interaction with molecular docking simulation. J Food Biochem 42(1):e12463
Durand DJ, Gordon Green B, O’Connell JF, Grant SK (1999) Peptide aldehyde inhibitors of bacterial peptide deformylases. Arch Biochem Biophys 367(2):297–302
Fremlin LJ, Piggott AM, Lacey E, Capon RJ (2009) Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus versicolor. J Nat Prod 72(4):666–670
Ghosh AK, Panda SK, Luyten W (2021) Anti-Vibrio and immune-enhancing activity of medicinal plants in shrimp: a comprehensive review. Fish Shellfish Immunol 117:192–210
Gonçalves VN, Campos LS, Melo IS, Pellizari VH, Rosa CA, Rosa LH (2013) Penicillium solitum: a mesophilic, psychrotolerant fungus present in marine sediments from Antarctica. Polar Biol 36(12):1823–1831
Gonçalves VN, Carvalho CR, Johann S, Mendes G, Alves TMA, Zani CL, Junior PAS, Murta SMF, Romanha AJ, Cantrell CL, Rosa CA, Rosa LH (2015) Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol 38(8):1143–1152
Grzela R, Nusbaum J, Fieulaine S, Lavecchia F, Desmadril M, Nhiri N, Dorsselaer AV, Cianferani S, Jacquet E, Meinnel T, Giglione C (2018) Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts. BBA-Proteins Proteom 1866(2):348–355
Handayani DP, Isnansetyo A, Istiqomah I, Jumina J (2022) Anti-Vibrio activity of Pseudoalteromonas xiamenensis STKMTI.2, a new potential vibriosis biocontrol bacterium in marine aquaculture. Aquac Res 53(5):1800–1813
Hussain FBM, Al-Khdhairawi AAQ, Kok Sing H, Muhammad Low AL, Anouar EH, Thomas NF, Alias SA, Manshoor N, Weber JFF (2020) Structure elucidation of the spiro-polyketide svalbardine B from the Arctic fungal endophyte Poaceicola sp. E1PB with support from extensive ESI-MSn interpretation. J Nat Prod 83(12):3493–3501
Ina-Salwany MY, Al-saari N, Mohamad A, Fathin-Amirah M, Mohd A, AmalM NA, Kasai H, Mino S, Sawabe T, Zamri-Saad M (2019) Vibriosis in fish: a review on disease development and prevention. J Aquatic Anim Health 31(1):3–22
Kang C, Shin Y, Jang S, Jung Y, So J (2016) Antimicrobial susceptibility of Vibrio alginolyticus isolated from oyster in Korea. Environ Sci Pollut Res 23(20):21106–21112
Li G, **e GS, Wang HL, Wan XY, Li XS, Shi CY, Wang ZY, Gong M, Li T, Wang P, Zhang QL, Huang J (2021) Characterization of a novel shrimp pathogen, Vibrio brasiliensis, isolated from pacific white shrimp, Penaeus Vannamei. J Fish Dis 44(10):1543–1552
Ma YM, Qiao K, Kong Y, Li MY, Guo LX, Miao Z, Fan C (2017) A new isoquinolone alkaloid from an endophytic fungus R22 of Nerium indicum. Nat Prod Res 31(8):951–958
Meinnel T (2000) Peptide deformylase of eukaryotic protists: a target for new antiparasitic agents? Parasitol Today 16(4):165–168
Meinnel T, Blanquet S (1993) Evidence that peptide deformylase and methionyl-tRNA(fMet) formyltransferase are encoded within the same operon in Escherichia coli. J Bacteriol 175(23):7737–7740
Meinnel T, Blanquet S (1994) Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNA(fMet) formyltransferase. J Bacteriol 176(23):7387–7390
Nakayama T, Ito E, Nomura N, Nomura N, Matsumura M (2006) Comparison of Vibrio harveyi strains isolated from shrimp farms and from culture collection in terms of toxicity and antibiotic resistance. FEMS Microbiol Lett 258(2):194–199
Pan CQ, Shi YT, Chen XG, Chen CTA, Tao XY, Wu B (2017) New compounds from a hydrothermal vent crab-associated fungus Aspergillus versicolor XZ-4. Org Biomol Chem 15(5):1155–1163
Ragunathan A, Malathi K, Anbarasu A (2018) MurB as a target in an alternative approach to tackle the Vibrio cholerae resistance using molecular docking and simulation study. J Cell Biochem 119(2):1726–1732
Raval IH, Labala RK, Raval KH, Chatterjee S, Haldar S (2021) Characterization of VopJ by modelling, docking and molecular dynamics simulation with reference to its role in infection of enteropathogen Vibrio parahaemolyticus. J Biomol Struct Dyn 39(5):1572–1578
Robinson CH (2001) Cold adaptation in Arctic and Antarctic fungi. New Phytol 151(2):341–353
Sasikala D, Jeyakanthan J, Srinivasan P (2016) Structural insights on identification of potential lead compounds targeting WbpP in Vibrio vulnificus through structure-based approaches. J Recept Signal Transduct Res 36(5):515–530
Schoch CL, Seifert KA, Huhndorf S, Robert V, AndréLevesque C, Spouge JL, Chen W, Consortium FB (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109(16):6241–6246
Shah M, Sun C, Sun Z, Zhang GJ, Che Q, Gu QQ, Zhu TJ, Li DH (2020) Antibacterial polyketides from Antarctica sponge-derived fungus Penicillium sp. HDN151272. Mar Drugs 18(2):71
Sun WG, Chen XT, Tong QY, Zhu HC, He Y, Lei L, Xue YB, Yao GM, Luo ZW, Wang JP, Li H, Zhang YH (2016) Novel small molecule 11β-HSD1 inhibitor from the endophytic fungus Penicillium commune. Sci Rep 6(1):26418
Wang LY, Li MJ, Lin YZ, Du SW, Liu ZY, Ju JH, Suzuki H, Sawada M, Umezawa K (2020) Inhibition of cellular inflammatory mediator production and amelioration of learning deficit in flies by deep sea Aspergillus-derived cyclopenin. J Antibiot 73(9):622–629
**n ZH, Fang YC, Zhu TJ, Duan L, Gu QQ, Zhu WM (2006) Antitumor components from sponge-derived fungus Penicillium auratiogriseum Sp-19. Chin J Mar Drugs 25(6):1–6
Yano Y, Hamano K, Satomi M, Tsutsuid I, Ma B, Aue-umneoy D (2014) Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food Control 38:30–36
Funding
This work was supported by the grants from the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 22KJB170010); the Scientific Research Foundation of Jiangsu Ocean University (No. KQ20047); the Program of Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening (No. HY202001); the Foundation from Jiangsu Province (No. JSSCBS20211307); the Program of MNR Key Laboratory of Coastal Salt Marsh Ecosystems and Resources (No. KLCSMERMNR2021107); the “Blue Project” of Jiangsu Higher Education Institutions of China, and the Priority Academic Program Development of Jiangsu Higher Education Institutions of China.
Author information
Authors and Affiliations
Contributions
JCG and JY carried out the isolation of the fungus and extraction of the secondary metabolites, performed the anti-Vibrio assay, structure characterization. BG and HFL performed the molecular docking and collaborated in the analysis of the obtained results. FLA provided the fungus and conceived this work. PW and DZ wrote the manuscript text. SG contributed to the data interpretation and the revision of the draft. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The detailed experiment procedures and the HRESIMS, NMR or UV spectra of compunds 1–7.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, J., Yang, J., Wang, P. et al. Anti-vibriosis bioactive molecules from Arctic Penicillium sp. Z2230. Bioresour. Bioprocess. 10, 11 (2023). https://doi.org/10.1186/s40643-023-00628-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-023-00628-5