Abstract
Sensitive quantification of microRNA (miRNA) plays a crucial role in early diagnosis and precise therapy of osteosarcoma. Herein, we build a label-free and sensitive miRNA quantification approach based on the activation of split-DNAzyme initiated primer exchange reaction (PER). Target miRNA cooperates the activation of split-DNAzyme with Mg2+ through assisting the assembly of DNAzyme to correct conformation, which enables the performance of PER-based nucleic acids amplification to produce a large amount of single-strand DNA (ssDNA) sequences. The G-quadruplexes (G4) in ssDNA sequences products bind with N-methyl mesoporphyrin IX (NMM) to form G4-NMM complex with the enhanced fluorescence respond. The results demonstrate that miRNA-21 can assist the activation of split-DNAzyme, and the active DNAzyme exhibits a high specificity and efficiency in inducing the subsequent PER. Based on the split-DNAzyme-assisted signal recycle and PER, the method eventually shows a high sensitivity and selectivity, providing a promising prospect for the for early stage tumor diagnosis and more precise tumor therapy.
Similar content being viewed by others
Introduction
Osteosarcoma is a common primary malignant bone tumor derived from mesenchymal tissue which has a high propensity for local invasion (Chen et al. 2021; Kansara et al. 2014). The lack of reliable biomarkers for the early diagnosis of osteosarcoma leads to a high distant metastasis rate (approximate 80%) and low 5-year survival rate of 10–20% (Kaste et al. 1999). Therefore, it is of great significance to develop reliable and effective biomarkers for early diagnosis of osteosarcoma, and providing valuable information for the long-term postoperative management. MicroRNA (miRNA) is a kind of small non-coding RNAs with the function of modulating gene expression, which is involved in diverse pathological processes, e.g., cancers (Acunzo et al. 2015; Hussen et al. 2021; Sempere et al. 2021). Recent researches implied the crucial roles of miRNA in regulating the proliferation and differentiation of osteosarcoma cells and hold the view that miRNA could be potentially applied for the diagnosis, prognosis and treatment of osteosarcoma (Sekar et al. 2019; Singh et al. 2021; Xu et al. 2021). For example, Hu reported that serum miRNA-21 was up-expressed as the T-stage osteosarcoma increased and demonstrated that miRNA-21 could inhibit the proliferation of osteosarcoma through targeting PTEN and regulating the TGF-1 signaling pathway (Hu et al. 2018). Therefore, it is in urgent demand to develop a sensitive yet reliable miRNA detection approach.
The conventional method for miRNA detection is qRT-PCR, which is the gold standard for clinical and laboratory trials (Hu et al. 2021; Saliminejad et al. 2019). The method contributes to the sensitive miRNA quantification. However, qRT-PCR requires thermal cycle, complicated primer design and lengthy incubation time, which limited their further applications (Ren et al. 2015). To combat the issue, a variety of isothermal signal amplification strategies have been proposed in recent years (Lobato and O'Sullivan 2018; Reid et al. 2018; Wong et al. 2018; Zhao et al. 2015), such as rolling circle amplification (RCA) and primer exchange reaction (PER) (Deng et al. 2020; Hollenstein 2018). Among them, PER has attracted abundant attention due to its unique capability of programmable autonomous synthesis of large amount of signal-strand DNA (ssDNA). Based on this, diverse PER-based miRNA detection approaches have been proposed. For example, Zhang built a tumor-associated biomarker initiated PER-based DNA machine and successfully applied it for miRNA imaging and gene therapy (Zhang et al. 2020). Despite that PER-based approaches exhibit a favorable detection performance, the limited sensitivity made it difficult to detect low-abundant miRNAs.
With this in mind, we depict here a label-free miRNA detection approach with an improved sensitivity by combining split-DNAzyme-based signal recycle with PER and using G-duplex for NMM-based signal generation. In this method, target miRNA-based activation of split-DNAzymes turns on subsequent PER through generating a nicking site in the loop section in deigned dumbbell structure probe. During the PER process, a large amount of ssDNA containing G-quadruplexes (G4) are produced and G4 sequences can bind with N-methyl mesoporphyrin IX (NMM) to form G4-NMM complex with the enhanced fluorescence respond that is correlated with the concentration of miRNA in the sensing system.
Material and experimental procedure
Materials and reagents
All oligonucleotides utilized in this research (listed in Additional file 1: Table S1) were brought from Shanghai Sangon Biological Engineering Technology (Shanghai, China) and purified by HPLC water. The Klenow fragment (KF) enzyme used for chain extension, deoxyribonucleotides mixture (dNTPs) and corresponding buffer solution were purchased from Sigma-Aldrich (Shanghai, China). NMM was obtained from Sangon Biological Engineering Technology (Shanghai, China).
Validation of the assembly of split-DNAzyme
Fluorescence assay
To test the assembly of split-DNAzyme, 10 μL synthesized split-DNAzyme-1 (20 μM), 10 μL synthesized split-DNAzyme-2 (20 μM) and 10 μL miRNA-21 (10 μM) were mixed with the buffer solution containing 100 mM Tris–HCl, 500 mM NaCl, 100 mM MgCl2 and 10 mM DTT. The mixture was firstly heated to 90 °C for 10 min and gradually cooled to room temperature within 30 min. 10 μL m sequences (10 μM) was then added in the mixture and incubated for 30 min at room temperature. The fluorescence signal of the mixture was then detected by the Hitachi F-4700 fluoro spectrophotometer (Tokyo, Japan).
PAGE analysis
Procedures of PAGE analysis were performed according to former reports. The samples were prepared as follows: Lane 1: miRNA (final concentration: 2 μM); Lane 2: mixture of split-DNAzyme-1 and split-DNAzyme-2 (2 μM, respectively); Lane 3: miRNA-21 + split-DNAzyme-2 + split-DNAzyme-1 (2 μM, respectively); Lane 4: m sequences (2 μM); and Lane 5: miRNA-21 + split-DNAzyme-2 + split-DNAzyme-1 + m sequences.
PER-based miRNA-21 detection
To verify the feasibility of the approach, 10 μL miRNA-21 (2 μM) was mixed with 10 μL synthesized split-DNAzyme-1 (2 μM) and 10 μL synthesized split-DNAzyme-2 (2 μM). The mixture was firstly heated to 90 °C for 5 min and gradually cooled to room temperature. 10 μL dumbbell probe (2 μM) was then added to the mixture and incubated for 15 min at room temperature. Afterward, 10 μL primer sequence (2 μM), 2 μL KF, 2 μL buffer solution and 6 μL DEPC water were added to the mixture. The mixture was incubated at 55 °C for 100 min and then incubated at 70 °C for 5 min to degrade KF enzyme. NMM was added in the mixture, and the fluorescence signal was detected after incubating for 10 min.
Results and discussion
Mechanism and design of the PER-based approach
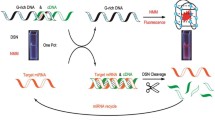
The working mechanism of the PER-based label-free miRNA detection approach is depicted in Fig. 1. Two split-DNAzyme sequences were first designed with three functional sections, respectively. The a sections in the two split-DNAzyme sequences could bind with miRNA-21; b section in the two sequences was complementary and could form a duplex only when a sections were hybridized with miRNA-21; the rest section in the two split-DNAzyme sequences could bind with loop section in the designed dumbbell probe. The dumbbell-shaped probe in the PER process was designed with two loop sections. In detail, the c sections could provide biding sites for primer; e section could transcribe G4 sequence; and d section assisted the formation of dumbbell probe. Based on the miRNA-21-assisted formation of correct secondary conformation of DNAzyme and Mg2+-assisted activation, split-DNAzyme-miRNA-21 complex bound with loop section in the dumbbell probe and generated a nicking site, exposing the recognizing section that could be identified by primer. When split-DNAzyme-miRNA-21 complex was discharged from a dumbbell probe, it could bind with a next dumbbell probe to form a signal cycle. With the aid of DNA polymerase (Klenow fragment, KF), a nascent ssDNA sequence was appended to the 3′ terminal of the primer, which was extended until the stand displacing extension reaction was haltered at the stop sequence on the dumbbell hairpin probe. The d section competed with copied d′ section and via the random walk process of three-way migration. As a result, the d section was released from dumbbell probe, and dumbbell probe was free to bind with another primer and induce a next PER. Therefore, a large amount of ssDNA sequences (e′ sequence) containing G4 sequences were generated, which could be identified by NMM to form G4-NMM complex with the enhanced fluorescence respond.
Feasibility of split-DNAzyme assembly and PER
Target miRNA-based assembly of split-DNAzyme determined the activation of DNAzyme and imitation of subsequent PER. The assembly of split-DNAzyme was firstly investigated through PAGE analysis. The Lane 1 in Fig. 2a was target miRNA-21, and Lane 2 was the complex of split-DNAzyme and miRNA-21. To test the cleavage activity of the active DNAzyme, a ssDNA sequence (m sequence) was synthesized to mimic the loop section in dumbbell probe. In Lane 3, a band was observed that moved faster than DNAzyme, indicating that m sequences were digested by the active DNAzyme. PAGE result of dumbbell probe-assisted formation of active DNAzyme is listed in Additional file 1: Fig. S1, indicating a same conclusion. Fluorescence assay was performed on FAM and BHQ-2 labeled m sequence, and the result showed a greatly enhanced signal when active DNAzyme existed (Fig. 2b). PER process was validated in the PAGE result in Fig. 2c. A hairpin probe was utilized to mimic the dumbbell probe which has a nicking site in the loop section. When the hairpin probe was mixed with primer, a new band appeared in Lane 3, moving slower than hairpin probe alone. After KF-based chain extension, a band between hairpin probe and primer appeared, which was considered the produced e′ sequences.
Investigation of assembly of split-DNAzyme and feasibility of PER. a PAGE analysis of the assembly of split-DNAzyme; b Fluorescence intensity of the m sequences when they were incubated with different combination of S1 (split-DNAzyme-1), S2 (split-DNAzyme-2), miRNA-21 and Mg2+. c PAGE analysis of the PER process
Analytical performance of the approach
For a better detection performance, several experimental parameters were optimized, including the length of b section in the two split-DNAzyme sequences, concentration of KF and incubation time. From the result in Additional file 1: Fig. S2, the length of b section was determined 5 nt; concentration of KF was 1 U/L; and the incubation time of the whole system was 1.5 h. In addition, the concentration ratio of split-DNAzyme for assembly was determined 1:1; the reaction temperature after adding the target miRNA-21 was optimized to 37 °C; and the concentration of NMM was determined 250 μM (Additional file 1: Fig. S3). Under the obtained optimized experimental conditions, we then studied the detection performance of the approach. To test the sensitivity of the approach, the approach was utilized to detect a series of samples containing synthesized miRNA-21 which was firstly diluted to different concentrations ranging from 10 fM to 1 nM. The detected fluorescence signal gradually increased when the concentration of miRNA in samples increased (Fig. 3a). The peak value of detected fluorescent signals was correlated with the logarithmic concentration of miRNA-21 in the sample. The correlation equation was determined Y = 1181*lgC + 10,135, with R2 of 0.9954 (Fig. 3b). In addition, the limit of detection (LOD) of the method was computed as 2.43 fM based on the general 3σ method. Selectivity of the approach was verified through detecting miRNA-21 and other miRNAs (miRNA-211, miRNA-155, Let-7a, 1 pM respectively). The result in Fig. 3c showed that fluorescence signal of the approach when detecting miRNA-21 was significantly high compared with control (without miRNA-21). When miRNA-211, miRNA-155 and Let-7a, the approach exhibited neglectable signal enhancements compared with control group, indicating a high selectivity of the approach. As a conventional approach, qRT-PCR is a gold-strand for miRNA quantification in clinical applications. Herein, we compared the detection performance of the established approach with qRT-PCR. The method and qRT-PCR were utilized to quantify 6 samples containing different concentrations of miRNA-21, respectively. The result in Fig. 3d showed a good correlation between the calculated miRNA concentrations by the established approach and by qRT-PCR, indicating a promising applicable potential of the approach.
Detection performance of the approach. a Fluorescence spectrum of the approach when detecting different concentrations of miRNA-21; b Correlation equation between peak value of fluorescence spectrum and the concentration of miRNA-21; c Fluorescence intensity of the approach when detecting different miRNAs; d Correlation of the calculated miRNA by the method and qRT-PCR
Clinical application of the approach
As a crucial biomarker, the expression level of miRNA-21 is up-regulated in osteosarcoma patients. Thus, we applied the approach to quantify miRNA-21 in MG-63 cell extraction and normal cell extraction (hOB cell). The result in Fig. 4a showed that miRNA-21 level in MG-63 cell extractive is much higher than that in the hOB cell extractive. The repeatability was verified through detecting 10 sample duplicates containing 100 pM miRNA, respectively. The result in Fig. 4b showed a coefficient of variation (CV) of 4.65%. Compared with former approaches, the established method exhibited a low LOD in a label-free manner which could meet the requirements of clinical practice (Additional file 1: Table S2).
Conclusion
In summary, we developed a label-free and sensitive miRNA detection approach. The approach was initiated by target miRNA-based assembly of split-DNAzyme to form active DNAzyme and to induce subsequent PER through generating a nicking site in the loop section in dumbbell probe. As a robust isothermal signal amplification tool, a large amount of ssDNA products containing G4 sequences were produced through PER, which could bind with NMM to form G4-NMM complex with enhanced signals. Based on the integration of DNAzyme-assisted signal recycle and PER, the method exhibited a wide detection range with a high sensitivity and eventually was successfully applied to distinguish miRNA-21 from a variety of miRNAs. Moreover, the capability of the approach to quantify miRNA-21 in MG-63 cell extractive and in serum samples indicates a promising prospect of the approach as a robust tool in clinical and laboratory applications.
Availability of data and materials
Almost all details of experimental data are presented in the article or additional file.
References
Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9.
Chen C, **e L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10.
Deng J, Li Y, Shi W, Liu R, Ma C, Shi C. Primer design strategy for denaturation bubble-mediated strand exchange amplification. Anal Biochem. 2020;593:113593.
Hollenstein M. DNA synthesis by primer exchange reaction cascades. ChemBioChem. 2018;19(5):422–4.
Hu X, Li L, Lu Y, Yu X, Chen H, Yin Q, Zhang Y. miRNA-21 inhibition inhibits osteosarcoma cell proliferation by targeting PTEN and regulating the TGF-beta1 signaling pathway. Oncol Lett. 2018;16(4):4337–42.
Hu C, Zhang L, Yang Z, Song Z, Zhang Q, He Y. Graphene oxide-based qRT-PCR assay enables the sensitive and specific detection of miRNAs for the screening of ovarian cancer. Anal Chim Acta. 2021;1174:338715.
Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: a signature for cancer progression. Biomed Pharmacother. 2021;138:111528.
Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–35.
Kaste SC, Pratt CB, Cain AM, Jones-Wallace DJ, Rao BN. Metastases detected at the time of diagnosis of primary pediatric extremity osteosarcoma at diagnosis: imaging features. Cancer. 1999;86(8):1602–8.
Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. Trends Analyt Chem. 2018;98:19–35.
Reid MS, Le XC, Zhang H. Exponential isothermal amplification of nucleic acids and assays for proteins, cells, small molecules, and enzyme activities: an EXPAR example. Angew Chem Int Ed Engl. 2018;57(37):11856–66.
Ren A, Dong Y, Tsoi H, Yu J. Detection of miRNA as non-invasive biomarkers of colorectal cancer. Int J Mol Sci. 2015;16(2):2810–23.
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–65.
Sekar D, Mani P, Biruntha M, Sivagurunathan P, Karthigeyan M. Dissecting the functional role of microRNA 21 in osteosarcoma. Cancer Gene Ther. 2019;26(7–8):179–82.
Sempere LF, Azmi AS, Moore A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA. 2021;12(6):e1662.
Singh A, Singh AK, Giri R, Kumar D, Sharma R, Valis M, Kuca K, Garg N. The role of microRNA-21 in the onset and progression of cancer. Future Med Chem. 2021;13(21):1885–906.
Wong YP, Othman S, Lau YL, Radu S, Chee HY. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J Appl Microbiol. 2018;124(3):626–43.
Xu Z, Chen L, Wang C, Zhang L, Xu W. MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4. Free Radic Res. 2021;55(11–12):1119–29.
Zhang J, He M, Nie C, He M, Pan Q, Liu C, Hu Y, Chen T, Chu X. Biomineralized metal-organic framework nanoparticles enable a primer exchange reaction-based DNA machine to work in living cells for imaging and gene therapy. Chem Sci. 2020;11(27):7092–101.
Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491–545.
Acknowledgements
The authors thank financial and technical support from Baoji Maternal and Child Health Hospital
Funding
No fund available.
Author information
Authors and Affiliations
Contributions
LX performed related experiments, analyzed data and wrote original manuscript. GM supported the research, designed the approach and assisted in writing of the manuscript. WH assisted in data analysis; XM assisted in data collection and processing; and JL and JZ checked spelling and grammar. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The manuscript does not contain clinical or trial studies on patients, humans or animals.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Table S1. Details of the sequences used in this research. Table S2. A brief comparison of the advantages and disadvantage of the method with former ones. Fig. S1. PAGE result of PAGE result of dumbbell probe assisted formation of active DNAzyme. Fig. S2. Optimization of experimental conditions. Fig. S3. Optimization of experimental conditions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, X., Wu, H., **ong, M. et al. Split-DNAzyme cooperating primer exchange reaction for sensitive miRNA detection. J Anal Sci Technol 13, 33 (2022). https://doi.org/10.1186/s40543-022-00343-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40543-022-00343-4