Abstract
Background
Rectal cancer is one of the most common malignancies. To predict the specific mortality risk of rectal cancer patients, we constructed a predictive nomogram based on a competing risk model.
Methods
The information on rectal cancer patients was extracted from the SEER database. Traditional survival analysis and specific death analysis were performed separately on the data.
Results
The present study included 23,680 patients, with 16,580 in the training set and 7100 in the validation set. The specific mortality rate calculated by the competing risk model was lower than that of the traditional survival analysis. Age, Marriage, Race, Sex, ICD-O-3Hist/Behav, Grade, AJCC stage, T stage, N stage, Surgery, Examined LN, RX SUMM-SURG OTH, Chemotherapy, CEA, Deposits, Regional nodes positive, Brain, Bone, Liver, Lung, Tumor size, and Malignant were independent influencing factors of specific death. The overall C statistic of the model in the training set was 0.821 (Se = 0.001), and the areas under the ROC curve for cancer-specific survival (CSS) at 1, 3, and 5 years were 0.842, 0.830, and 0.812, respectively. The overall C statistic of the model in the validation set was 0.829 (Se = 0.002), and the areas under the ROC curve for CSS at 1, 3, and 5 years were 0.851, 0.836, and 0.813, respectively.
Conclusions
The predictive nomogram based on a competing risk model for time-specific mortality in patients with rectal cancer has very desirable accuracy. Thus, the application of the predictive nomogram in clinical practice can help physicians make clinical decisions and follow-up strategies.
Similar content being viewed by others
Background
Rectal cancer (RC) is a type of cancer that occurs at the dentate line and colon-sigmoideum junctions [1]. Colorectal cancer is the fourth deadliest cancer in the world [2], and RC represents 40% of colorectal cancer cases. In the United States, there are 44,850 new RC cases (26,650 in men; 18,200 in women) in 2022 and 46,050 new RC cases (27,440 in men, 18,610 in women) in 2023, suggesting an increasing incidence [3]. Furthermore, the morbidity and mortality of RC are projected to rise dramatically by 2035 [4]. This cancer may be caused by several controllable risk factors, such as sedentary work [OR = 1.08 (95%CI: 1.00–1.16)] [5], male obesity [RR = 1.12 (95%CI: 1.09–1.16)] [6], red meat intake [RR = 1.22 (95% CI: 1.01–1.46)] [7], meat products [RR = 1.26 (95% CI = 1.09–1.45)] [7], and high alcohol intake [OR = 1.25 (95%CI: 1.11–1.40)] [8].
The distal third of the rectum is drained by systemic veins instead of the portal venous system, which increases the likelihood of direct lung metastases [9]. RC penetrates the pelvis more easily than colon cancer, with a higher risk of lymph node metastasis in each T stage, complex anatomical structures, difficult-to-operate surgery, and a high postoperative recurrence rate. Currently, treatments for RC include local or extensive surgery, preoperative radiotherapy and systemic therapy, local ablation of metastases, palliative chemotherapy, targeted therapy, and immunotherapy [10]. The selection of treatments is based on the tumor’s clinicopathological characteristics, such as the degree of differentiation and the site of metastasis [11, 12]. RC survival is closely related to clinical decisions based on staging information. The 5-year disease-free survival (DFS) and overall survival (OS) of patients with RC were 70.1% (95%CI: 60.7%–73.0%) and 76.2% (95%CI: 73.8%–78.5%) with open surgery, and 72.2% (95%CI: 69.4%–74.8%) and 72.7% (95%CI: 69.8%–75.3%) with laparoscopic surgery, respectively [13]. Compared with long-course chemoradiotherapy, both total neoadjuvant therapy with long-course chemoradiotherapy [OR = 1.78 (95%CI: 1.43–2.26)] and total neoadjuvant therapy with short-course radiotherapy [OR = 1.75(95%CI: 1.23–2.50)] improved the pathological complete response rate; neoadjuvant chemotherapy increased DFS, but did not significantly prolong OS [14]. At present, TNM staging is the primary reference in clinical practice. Other potential prognostic factors, such as age, sex, marriage, and region, are also strongly associated with prognosis [15]. Therefore, it is necessary to explore a more comprehensive prediction method. Clinical predictive models are more personalized approaches tailored to these factors.
The specific mortality risk is more valuable than the traditional mortality risk in clinical practice due to some unforeseeable mortality events. Specifically speaking, the causes of death in RC patients are diverse, and a large number of mortality events are not caused by RC, as RC patients are often in poor functional status or accompanied by co-morbidities [16]. These deaths are unforeseeable when patients die of other diseases, such as cardiovascular disease. In addition, as competing risks often occur among the diverse risk factors for death in patients with RC, traditional survival analysis methods may ignore the occurrence of main concern events, resulting in overestimated results [17]. Therefore, traditional survival analysis methods are unsuitable for processing data containing competing outcomes. A competing risk model is more suitable for dealing with data containing competing events and can produce more accurate results [17].
Since most patients with RC have a long survival period, identifying the risk of specific death early, in the case of competing events, is necessary to facilitate the development of specific prognostic rehabilitation programs and follow-up plans, thereby reducing the risk of specific death in patients. Although various prognostic models, especially based on radiomics, have been established in recent years, predictive models in clinical practice should be highly interpretable and applicable. Therefore, we conducted this study based on the SEER database to construct an interpretable nomogram for early predicting the risk of RC-specific mortality.
Methods
Data sources and extractions
The SEER database, which comprises 18 cancer registries, is the National Cancer Institute's collection of demographic statistics on all cancers diagnosed in representative geographic regions and subpopulations. We collected patient clinicopathological data, demographic information, clinicopathological information, and follow-up data from the database. The patient data were de-identified because the SEER database is publicly available. In addition, ethical approval and informed consent are not required for our study. Our methodology strictly follows the rules of the SEER database.
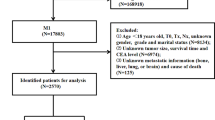
We selected the “Incidence SEER Research Plus Data, 18 Registries, Nov 2020 Sub (2000–2018)” database from 2010 to 2015. According to the disease name of RC, we collected the clinicopathological information of all RC patients, including Patient ID, Age, Marriage, Race, Sex, Primary Site- labeled, ICD-O-3Hist/Behav, Grade, Laterality, AJCC stage, AJCC Tstage, AJCC N stage, AJCC M stage, Surgery, Examined LN, RX SUMM-SURG OTH REG, Radiation recode, Chemotherapy, Preoperative CEA, Pleural Effusion Recode, Tumor deposits, Regional nodes positive, Brain, Bone, Liver, Lung, CS Tumor size, Malignant tumor numbers, cause of death classification, Survival months, COD to site rec KM. Exclusion criteria were as follows: (1) unclear modeling variables NA, Blanks, unknown TNM stage (TX, NX); (2) unknown tumor size; (3) unknown surgical method; (4) survival time < 1 month or unknown (5) Age ≤ 18.
Construction and verification of competitive risk model
We developed a competing risk model by defining deaths from unknown causes or survival cases in RC as censoring events, deaths from specific causes as interest events, and deaths from other causes as competing events. Before building the model, we randomly split the data by 70% and 30% into training and validation sets. A uniform random number distributed between 0 and 1 was assigned to each case, and then these cases were ranked according to their random number. The cases with a random number < 70 percentile were assigned to the training set, and the rest of the cases were assigned to the validation set. Univariate and multivariate analyses were performed in the training set to screen the independent influencing factors of RC, and RC-specific death nomograms were constructed according to the independent influencing factors. The concordance index (C-index) and the area under the receiver operating curve (AUC) were used to measure the accuracy of the prediction nomogram, and the calibration curve was used to measure the calibration degree of the prediction nomogram.
Statistical analysis
Continuous variables were tested for normal distribution using Shapiro–Wilk test. Normal distributed continuous variables were expressed as mean ± standard deviation (SD), and an independent samples t-test was used to compare the means of the two groups. For continuous variables with skewed distribution, the Mann–Whitney U test was performed for comparisons between groups. Count data were expressed as frequency (%); and the chi-square test was used for comparison between multiple groups. A P < 0.05 indicated a statistically significant difference. R 4.2.0 was used for modeling, while SPSS 2.3.0 was used for statistical analysis.
Results
Patient characteristics
A total of 23,680 RC patients were included in the analysis. Patients were randomly assigned to the training cohort (n = 16,580) and the validation cohort (n = 7100). In the training cohort, the mean age of patients was 63.02 ± 13.38 years; the number of deaths was 6470; the number of specific deaths was 4602; and the mean follow-up time was 52.35 ± 27.20 years. In the validation cohort, the mean age of patients was 62.90 ± 13.41 years; the number of deaths was 9174; the number of specific deaths was 6537; and the mean follow-up time was 52.53 ± 27.77 years. It can be seen that the distribution of the two groups is even, and the rest of the information is shown in Table 1.
Competing risk model for univariate and multivariate analysis
The variables in the present study included Age, Marriage, Race, Sex, ICD-O-3Hist/Behav, Grade, AJCC stage, T stage, N stage, Surgery, Examined LN, RX SUMM-SURG OTH, Chemotherapy, CEA, Deposits, Regional nodes in single factor Positive, Brain, Bone, Liver, Lung, Tumor size, and Malignant variables. Both univariate and multivariate analyses did not include the M stage because it overlapped with the AJCC stage. The variable Radiation (P > 0.05) was excluded from the multivariate analysis, whereas other variables in the single factor analysis were included in the multivariate analysis, with P < 0.05 (Table 2). The results showed that these variables were all independent influencing factors for RC. Accordingly, we constructed a competing risk model based on these variables.
Validation of predictive nomogram
Specific mortality prediction nomograms were drawn according to the competing risk model described above (Fig. 1). In the training set, the overall C statistic was 0.821 (Se 0.001), and the 1-, 3-, and 5-year AUC for predicting the specific mortality was 0.842 (95% CI 0.830–0.854), 0.830 (95% CI 0.822–0.838), 0.812 (95% CI 0.803–0.820), respectively. The results show that the model has a good identification ability (Fig. 2A). In addition, the model's calibration curve (Fig. 3A) shows that the 1-, 3- and 5-year curves are consistent with the ideal curve, indicating that the predicted curve diagram has good accuracy and calibration.
At the same time, in the internal validation cohort, the overall C statistic was 0.829 (Se 0.002); the 1-, 3-, and 5-year AUC (Fig. 2B) was 0.851 (95% CI 0.832–0.869), 0.836 (95% CI 0.824–0.849), and 0.813 (95% CI 0.800–0.826). The results also show that the model has good identification power. According to the calibration curve of the model (Fig. 3B), the 1-, 3- and 5-year curves are consistent with the ideal curve, indicating that the predicted curve diagram has good accuracy and calibration.
Specific mortality estimation
We compared the 1- to 9-year specific mortality calculated by traditional survival analysis and the competing risk model. The results revealed that the competing risk model showed lower specific mortality than traditional survival analysis. For example, the 1-year mortality rate was 0.08 in the traditional survival analysis and 0.07 in the specific survival analysis. Their gap grows as the survival duration increases between 1 and 9 years. Therefore, mortality from other causes could not be ignored (Table 3).
Discussion
Based on univariate and multivariate analyses, we included the independent influencing factors of RC-specific death, including Age, Marriage, Race, Sex, ICD-O-3Hist/Behav, Grade, AJCC stage, T stage, N stage, Surgery, Examined LN, RX SUMM-SURG OTH, Chemotherapy, CEA, Deposits, Regional nodes positive, Brain, Bone, Liver, Lung, Tumor size, and Malignant. These variables were combined to construct a competing risk model for RC and a predictive nomogram, which have excellent accuracy. The overall C statistic was 0.821 (Se 0.001) in the model training cohort and 0.829 (Se 0.002) in the internal validation cohort.
Currently, there are various RC prediction nomograms based on clinical data [ The data that support the findings of this study are available from the corresponding author upon reasonable request. Rectal cancer Cancer-specific survival Carcinoembryonic antigen Tumor deposits Lymph node metastasis Reactive oxygen species Wilkinson N. Management of rectal cancer. Surg Clin N Am. 2020;100(3):615–28. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (London, England). 2019;394(10207):1467–80. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer J Clin. 2023;73(1):17–48. Araghi M, Soerjomataram I, Jenkins M, Brierley J, Morris E, Bray F, Arnold M. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer. 2019;144(12):2992–3000. Lee J, Lee J, Ahn J, Lee DW, Kim HR, Kang MY. Association of sedentary work with colon and rectal cancer: systematic review and meta-analysis. Occup Environ Med. 2022;79(4):277–86. Larsson SC, Wolk A. Obesity and colon and rectal cancer risk: a meta-analysis of prospective studies. Am J Clin Nutr. 2007;86(3):556–65. Farvid MS, Sidahmed E, Spence ND, Mante Angua K, Rosner BA, Barnett JB. Consumption of red meat and processed meat and cancer incidence: a systematic review and meta-analysis of prospective studies. Eur J Epidemiol. 2021;36(9):937–51. McNabb S, Harrison TA, Albanes D, Berndt SI, Brenner H, Caan BJ, Campbell PT, Cao Y, Chang-Claude J, Chan A, et al. Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. Int J Cancer. 2020;146(3):861–73. Hou J, Zhang Y, Zhu Z. Gene heterogeneity in metastasis of colorectal cancer to the lung. Semin Cell Dev Biol. 2017;64:58–64. Kosinski L, Habr-Gama A, Ludwig K, Perez R. Shifting concepts in rectal cancer management: a review of contemporary primary rectal cancer treatment strategies. Cancer J Clin. 2012;62(3):173–202. O’Cathail SM, Smith T, Owens R, Zeniou A, Tsang Y, Holyoake DLP, Murray L, Harrison M, Hawkins MA. Superior outcomes of nodal metastases compared to visceral sites in oligometastatic colorectal cancer treated with stereotactic ablative radiotherapy. Radiother Oncol. 2020;151:280–6. Lee SA, Kwon HC, Park MA, Jung CK, Kim SH, Park KJ, Choi HJ, Lee HS, Roh MS, Kim JS, et al. Impact of the new AJCC staging system and adjuvant treatment in rectal cancer. Cancer Res Treat. 2004;36(2):121–7. Kong M, Chen H, Shan K, Sheng H, Li L. Comparison of survival among adults with rectal cancer who have undergone laparoscopic vs open surgery: a meta-analysis. JAMA Netw Open. 2022;5(5): e2210861. Wu Q, Zhou J, Huang J, Deng X, Li C, Meng W, He Y, Wang Z. Total neoadjuvant therapy versus chemoradiotherapy for locally advanced rectal cancer: Bayesian network meta-analysis. Br J Surg. 2023;110(7):784–96. Hases L, Ibrahim A, Chen X, Liu Y, Hartman J, Williams C. The importance of sex in the discovery of colorectal cancer prognostic biomarkers. Int J Mol Sci. 2021;22(3):56. Rutten HJ, den Dulk M, Lemmens VE, van de Velde CJ, Marijnen CA. Controversies of total mesorectal excision for rectal cancer in elderly patients. Lancet Oncol. 2008;9(5):494–501. Austin PC, Lee DS, Fine JP. Introduction to the analysis of survival data in the presence of competing risks. Circulation. 2016;133(6):601–9. Shen L, van Soest J, Wang J, Yu J, Hu W, Gong YU, Valentini V, **ao Y, Dekker A, Zhang Z. Validation of a rectal cancer outcome prediction model with a cohort of Chinese patients. Oncotarget. 2015;6(35):38327–35. Yu H, Huang T, Feng B, Lyu J. Deep-learning model for predicting the survival of rectal adenocarcinoma patients based on a surveillance, epidemiology, and end results analysis. BMC Cancer. 2022;22(1):210. Brunner W, Widmann B, Marti L, Tarantino I, Schmied BM, Warschkow R. Predictors for regional lymph node metastasis in T1 rectal cancer: a population-based SEER analysis. Surg Endosc. 2016;30(10):4405–15. Fan S, Li T, Zhou P, Peng Q, Zhu Y. Development and validation of nomogram combining serum biomarker for predicting survival in patients with resected rectal cancer. Biosci Rep. 2019;39(11):78. Diao JD, Wu CJ, Cui HX, Bu MW, Yue D, Wang X, Liu YL, Yang YJ. Nomogram predicting overall survival of rectal squamous cell carcinomas patients based on the SEER database: A population-based STROBE cohort study. Medicine. 2019;98(46): e17916. Lau B, Cole SR, Gange SJ. Competing risk regression models for epidemiologic data. Am J Epidemiol. 2009;170(2):244–56. Wang Y, Wu J, He H, Ma H, Hu L, Wen J, Lyu J. Nomogram predicting cancer-specific mortality in early-onset rectal cancer: a competing risk analysis. Int J Colorectal Dis. 2020;35(5):795–804. Lambregts DMJ, Bogveradze N, Blomqvist LK, Fokas E, Garcia-Aguilar J, Glimelius B, Gollub MJ, Konishi T, Marijnen CAM, Nagtegaal ID, et al. Current controversies in TNM for the radiological staging of rectal cancer and how to deal with them: results of a global online survey and multidisciplinary expert consensus. Eur Radiol. 2022;32(7):4991–5003. Cao Y, Deng S, Yan L, Gu J, Mao F, Xue Y, Zheng C, Yang M, Liu H, Liu L, et al. An oxidative stress index-based score for prognostic prediction in colorectal cancer patients undergoing surgery. Oxid Med Cell Longev. 2021;2021:6693707. Hutter RV. At last–worldwide agreement on the staging of cancer. Arch Surg. 1987;122(11):1235–9. Quirke P, Williams GT, Ectors N, Ensari A, Piard F, Nagtegaal I. The future of the TNM staging system in colorectal cancer: time for a debate? Lancet Oncol. 2007;8(7):651–7. **ang W, Lv Q, Shi H, **e B, Gao L. Aptamer-based biosensor for detecting carcinoembryonic antigen. Talanta. 2020;214: 120716. Margalit O, Mamtani R, Yang YX, Reiss KA, Golan T, Halpern N, Aderka D, Giantonio B, Shacham-Shmueli E, Boursi B. Assessing the prognostic value of carcinoembryonic antigen levels in stage I and II colon cancer. Eur J Cancer (Oxford, England, 1990). 2018;94:1–5. Konishi T, Shimada Y, Hsu M, Tufts L, Jimenez-Rodriguez R, Cercek A, Yaeger R, Saltz L, Smith JJ, Nash GM, et al. Association of preoperative and postoperative serum carcinoembryonic antigen and colon cancer outcome. JAMA Oncol. 2018;4(3):309–15. Alese OB, Zhou W, Jiang R, Zakka K, Huang Z, Okoli C, Shaib WL, Akce M, Diab M, Wu C, et al. Predictive and prognostic effects of primary tumor size on colorectal cancer survival. Front Oncol. 2021;11: 728076. Cai D, Huang ZH, Yu HC, Wang XL, Bai LL, Tang GN, Peng SY, Li YJ, Huang MJ, Cao GW, et al. Prognostic value of preoperative carcinoembryonic antigen/tumor size in rectal cancer. World J Gastroenterol. 2019;25(33):4945–58. Cohen R, Shi Q, Meyers J, ** Z, Svrcek M, Fuchs C, Couture F, Kuebler P, Ciombor KK, Bendell J, et al. Combining tumor deposits with the number of lymph node metastases to improve the prognostic accuracy in stage III colon cancer: a post hoc analysis of the CALGB/SWOG 80702 phase III study (Alliance)(☆). Ann Oncol. 2021;32(10):1267–75. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Garrido-Laguna I, et al. NCCN Guidelines Insights: Rectal Cancer, Version 6.2020. J Natl Comprehen Cancer Netw. 2020;18(7):806–15. Zheng H, Zhang J, Liu Y, Wang X. Prognostic value of tumor deposits in locally advanced rectal cancer: a retrospective study with propensity score matching. Int J Clin Oncol. 2021;26(6):1109–19. Delattre JF, Selcen Oguz Erdogan A, Cohen R, Shi Q, Emile JF, Taieb J, Tabernero J, André T, Meyerhardt JA, Nagtegaal ID, et al. A comprehensive overview of tumour deposits in colorectal cancer: Towards a next TNM classification. Cancer Treatment Rev. 2022;103:102325. Bruinvels DJ, Stiggelbout AM, Kievit J, van Houwelingen HC, Habbema JD, van de Velde CJ. Follow-up of patients with colorectal cancer. A meta-analysis. Ann Surg. 1994;219(2):174–82. Nicholson BD, Shinkins B, Pathiraja I, Roberts NW, James TJ, Mallett S, Perera R, Primrose JN, Mant D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev. 2015;2015(12):CD011134. Lord AC, Graham Martínez C, D’Souza N, Pucher PH, Brown G, Nagtegaal ID. The significance of tumour deposits in rectal cancer after neoadjuvant therapy: a systematic review and meta-analysis. Eur J Cancer. 2019;122:1–8. Genua F, Raghunathan V, Jenab M, Gallagher WM, Hughes DJ. The role of gut barrier dysfunction and microbiome dysbiosis in colorectal cancer development. Front Oncol. 2021;11: 626349. Janion K, Szczepańska E, Nowakowska-Zajdel E, Strzelczyk J, Copija A. Selected oxidative stress markers in colorectal cancer patients in relation to primary tumor location-a preliminary research. Medicina. 2020;56:2. Pappas-Gogos G, Tellis C, Lasithiotakis K, Tselepis AD, Tsimogiannis K, Tsimoyiannis E, Chalkiadakis G, Chrysos E. Oxidative stress markers in laparoscopic versus open colectomy for cancer: a double-blind randomized study. Surg Endosc. 2013;27(7):2357–65. Guo C, Li X, Wang R, Yu J, Ye M, Mao L, Zhang S, Zheng S. Association between oxidative dna damage and risk of colorectal cancer: sensitive determination of urinary 8-hydroxy-2’-deoxyguanosine by UPLC-MS/MS Analysis. Sci Rep. 2016;6:32581. Hou M, Sun JH. Emerging applications of radiomics in rectal cancer: State of the art and future perspectives. World J Gastroenterol. 2021;27(25):3802–14. Stanzione A, Verde F, Romeo V, Boccadifuoco F, Mainenti PP, Maurea S. Radiomics and machine learning applications in rectal cancer: Current update and future perspectives. World J Gastroenterol. 2021;27(32):5306–21. Ferro M, Falagario UG, Barone B, Maggi M, Crocetto F, Busetto GM, Giudice FD, Terracciano D, Lucarelli G, Lasorsa F, et al. Artificial intelligence in the advanced diagnosis of bladder cancer-comprehensive literature review and future advancement. Diagnostics. 2023;13(13):89. Ferro M, Crocetto F, Barone B, Del Giudice F, Maggi M, Lucarelli G, Busetto GM, Autorino R, Marchioni M, Cantiello F, et al. Artificial intelligence and radiomics in evaluation of kidney lesions: a comprehensive literature review. Ther Adv Urol. 2023;15:17562872231164804. Bedrikovetski S, Dudi-Venkata NN, Kroon HM, Seow W, Vather R, Carneiro G, Moore JW, Sammour T. Artificial intelligence for pre-operative lymph node staging in colorectal cancer: a systematic review and meta-analysis. BMC Cancer. 2021;21(1):1058. Di Re AM, Sun Y, Sundaresan P, Hau E, Toh JWT, Gee H, Or M, Haworth A. MRI radiomics in the prediction of therapeutic response to neoadjuvant therapy for locoregionally advanced rectal cancer: a systematic review. Expert Rev Anticancer Ther. 2021;21(4):425–49. Bourbonne V, Schick U, Pradier O, Visvikis D, Metges JP, Badic B. Radiomics approaches for the prediction of pathological complete response after neoadjuvant treatment in locally advanced rectal cancer: ready for prime time? Cancers. 2023;15(2):432. Staal FCR, van der Reijd DJ, Taghavi M, Lambregts DMJ, Beets-Tan RGH, Maas M. Radiomics for the prediction of treatment outcome and survival in patients with colorectal cancer: a systematic review. Clin Colorectal Cancer. 2021;20(1):52–71. We would like to thank the researchers and study participants for their contributions. This work was supported by the Henan Provincial Government-Health and Family Planning Commission Research Science Innovative Talents Project (51282). Conceptualization: RH, SD; Methodology: RH, SD; Formal analysis and investigation: RH, XZ, XL; Writing original draft preparation: RH; Writing review and editing: RH, SD; Funding acquisition: SD, XZ; Resources: RH, SD, XZ, XL; Supervision: SD; and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. Not applicable. Not applicable. The authors declare that they have no conflicts of interest to report regarding the present study. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data. Hu, R., Li, X., Zhou, X. et al. Development and validation of a competitive risk model in patients with rectal cancer: based on SEER database.
Eur J Med Res 28, 362 (2023). https://doi.org/10.1186/s40001-023-01357-3 Received: Accepted: Published: DOI: https://doi.org/10.1186/s40001-023-01357-3Availability of data and materials
Abbreviations
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Consent for publication
Competing interests
Additional information
Publisher's Note
Rights and permissions
About this article
Cite this article
Keywords