Abstract
Objective
This systematic review aims to outline the use of population and disease registries for clinical trial pre-screening.
Materials and methods
The search was conducted in the time period of January 2014 to December 2022 in three databases: MEDLINE, Embase, and Web of Science Core Collection. References were screened using the Rayyan software, firstly based on titles and abstracts only, and secondly through full text review. Quality of the included studies was assessed using the List of Included Studies and quality Assurance in Review tool, enabling inclusion of publications of only moderate to high quality.
Results
The search originally identified 1430 citations, but only 24 studies were included, reporting the use of population and/or disease registries for trial pre-screening. Nine disease domains were represented, with 54% of studies using registries based in the USA, and 62.5% of the studies using national registries. Half of the studies reported usage for drug trials, and over 478,679 patients were identified through registries in this review. Main advantages of the pre-screening methodology were reduced financial burden and time reduction.
Discussion and conclusion
The use of registries for trial pre-screening increases reproducibility of the pre-screening process across trials and sites, allowing for implementation and improvement of a quality assurance process. Pre-screening strategies seem under-reported, and we encourage more trials to use and describe their pre-screening processes, as there is a need for standardized methodological guidelines.
Similar content being viewed by others
Introduction
Clinical trials are essential in allowing the scientific transition from basic research to clinical practice, whether the trials are about drug development or other types of non-drug interventions [1]. Clinical trial protocols are the trial documents of reference, detailing every step for participants enrolled in the trial. One of the critical protocol sections, is the list of eligibility criteria [2]—if this list is clinical trial specific, it will often include recurring criteria within the same field or disease area, with trial specific cut-off differences [2]. For example, when conducting a trial for the neurodegenerative disease amyotrophic lateral sclerosis (ALS), it will in the majority of cases include vital capacity (VC) measurement as a trial eligibility criteria. Some ALS trials only include patients with a VC above 50% (NCT05633459), while others ask for patients with VC equal or superior to 65% (national clinical trial NCT, NCT04248465). Enrolling the right participants in a clinical trial is essential as it (1) could allow for a personalized medicine approach [3,4,5,6], (2) might be the only option to access drugs in development for patients suffering from diseases with no cures [7,8,9,10], (3) should ensure that motivated participants complete the entire study without drop** out, and thus ultimately maximize patient retention [11,12,13,14], and (4) in the end ensures good quality clinical trial data [15]. However, trial enrollment can also be challenging, especially to efficiently identify the above mentioned potentially eligible candidates during the pre-screening process [16,17,18]. This requires the identification of participants meeting the most stringent criteria and ultimately highly likely to be successful during the screening process. The pre-screening procedure is crucial as it decreases the screen failure rate, which drastically varies between trials across disease areas and countries [19,20,21,22]. Considering that screen failures are associated with participant burden while also negatively impacting the study budget, there is a need to develop clinical trial recruitment strategies targeting these aspects [23,24,25,26].
Typically, trial pre-screening is staff-bound with a designated staff member in charge of the pre-screening process. Research teams usually have team specific pre-screening processes, as there are no national consensus, guidelines nor universal standard operating procedures (SOPs) on how to conduct the pre-screening for clinical trials. A typical pre-screening process may include an internal check of the hospital medical journals in paper format, a review of electronic medical journals, a review of medical journals sent via traditional mail in case of a referral, direct emails from patients emailing a research team, and more [27,28,29,30]. This creates a pool of information derived from several sources, with no standardized system assuring quality and replicability, not allowing an audit trail for quality assurance and control, and overall creating inequitable trial access for patients [31]. Clinical trial eligibility criteria will often include both demographic data and disease specific information. This information is captured in most disease registries/population registries/patient registries, and there is an growing interest in using such registries for pre-screening due to the high quality and easy accessible data [32,33,34,35]. Such disease registries/population registries/patient registries are to be distinguished from other types of databases such as electronic health records (EHR). For the purpose of this review, we will use the term patient registry when referring to a specific database aiming to capture data on all patients from a patient population in a specific site, state, or country [32, 36, 37].
The usage of population and disease registries for trial pre-screening has previously been investigated through a literature review of the period 2004–2013, reporting limited registry use for clinical trial pre-screening, but advocating for a more systematic usage as this was deemed an efficient method [38]. The combined use of registries and medical record data has been described as optimizing trial recruitment [23], and we have since 2013 observed an explosion of clinical trials in many different fields as reported by the International Clinical Trial Registry Platform (ICTRP) of the World Health Organization (WHO) [39]. The ICTRP collects trial registration information from different databases such as Clinicaltrial.gov and reported 34,291 clinical trials in 2013 versus 59,964 clinical trials in 2021. The increasing number of clinical trials globally highlights the need for efficient and equitable pre-screening processes, but also for an updated review considering the last review on this topic was not conducted with a systematic methodology [38].
In this systematic review, we characterized the use of population and disease registries as a pre-screening tool for clinical trials not discriminating between drug and non-drug trials. We included publications published between January 2014 and December 2022, as a non-systematic review covered the 2004 to 2013 timeframe. We aimed to describe the type of registries used, disease areas, type of clinical trials linked to the registry-based pre-screening, and potential assets the method brought to the pre-screening process.
Methods
Inclusion and exclusion criteria
Citations and references obtained from the search were screened using the Rayyan software and our set of inclusion and exclusion criteria are listed in Table 1. Eligible studies had to be in English, from peer-reviewed journals, reporting the use of population/patient/disease registries for trial pre-screening. Included studies also needed to be set in trials on patients and not on healthy individuals. Studies had to have been published in our targeted window between January 2014 and December 2022, and abstracts had to be available for review. Finally, we included studies of high to moderate quality, as evaluated through the List of Included Studies and quality Assurance in Review (LISA-R) tool. Since there was no standardized tool to judge the quality of the included studies, we developed a quality assurance tool, the LISA-R. This quality assurance checklist was developed using guidance provided on the Parsifal platform for systematic reviews, a platform providing support for researchers conducting reviews and wishing to establish new quality assurance tools. The tool consists of 11 items in which each item was judged on a two-level scale (yes/no) (LISA-R blank tool available in Supplementary material 2). For each “yes”, one point was attributed, giving a scale range from 0 to 11. An overall score > 8 was interpreted as high quality, 6–8 moderate quality, and < 6 low quality.
Search and selection strategy
The study protocol was registered in PROSPERO with the identification number CRD42023433968 and followed the PRISMA requirements [40]. A literature search was performed in the following databases: MEDLINE, Embase, and Web of Science Core Collection. The last search was conducted on June 22, 2023.
The search strategy was developed in MEDLINE (Ovid) in collaboration with librarians at the Karolinska Institutet University Library. For each search concept, medical subject headings (MeSH-terms) and free text terms were identified (Supplementary material 1). The search was then translated, with Polyglot Search Translator used for the translation of the controlled vocabulary [41], into the other databases.
Language restriction was made to English and the search was limited to years 2014–2022 as a previous non-systematic review covered the 2004–2013 period [38]. De-duplication was done using the method described by Bramer et al. [1]. One final step was added to compare digital object identifiers to finalize de-duplication. The full search strategies for all databases are available in supplementary material (Supplementary material 1). The review of papers was conducted by two of the authors (JF and LA) independently and then cross-checked. A third author (CI) was asked to solve selection conflicts if they arose, by setting-up a meeting where JF and LA could expose their process and CI could make the final decision. A first review process (phase 1) was done based on titles and abstracts only, while the second review was of full texts (phase 2). Only the publications of moderate and high quality as per the LISA-R tool were included in the final search (phase 3).
Data extraction
Data extraction was conducted by two of the authors (JF and LA) reading the full texts and summarizing information in table format through an excel form. This data extraction form was created for the sole purpose of this systematic review. The extraction form included the information we wished to extract from the included studies: trial type (drug trial versus non-drug trial), clinical trial name, NCT number, registry name and scope, patient population, and age. In order to specifically look into enrollment and pre-screening rates, we extracted the number of patients identified through the registries, number of patients eligible for the trial in question, and number of patients enrolled in the clinical trial. Different enrollment rates were calculated when possible and represented by percentages: (1) comparing the number of patient enrolled to the number identified in the pre-screening process and (2) comparing the number of patient enrolled to number of patients actually eligible after screening.
Results
Review process
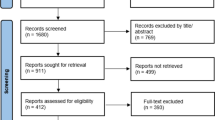
A total of 1430 citations were identified through the literature search. Out of them, 1369 were excluded based on titles and abstract review as they did not meet inclusion criteria (Table 1). One citation was excluded as a duplicate (Fig. 1). The 60 remaining publications were reviewed by reading the full text and 35 publications were subsequently excluded as they did not meet inclusion criteria. The remaining 25 publications were assessed using the LISA-R tool and the articles of low quality were excluded, ending up with 24 included papers (Supplementary material 3a and b). The list of excluded papers is available upon request.
Included articles—descriptive characteristics
Out of the 24 articles included and reporting the use of a population/disease registry for a clinical trial pre-screening [42,43,44,45,46,47,48,49,50,51,52,53,54,38, 74,75,76,77,78,79]. Our search highlighted that such recruitment registries seem to be extensively used in Alzheimer and dementia research [80,81,82,83,84,85,86,87]. This could explain why patient registries may surprisingly not be the first in line of use for trial pre-screening, as “recruitment registries” are blooming to support different trials. However, recruitment registries should be carefully considered as they bring ethical concerns. Indeed, they can lead to consenting patients already enrolled in other trials, or having to deal with changes in patient’s disease status not being updated [88].
In terms of disease areas, 11 of the 24 included studies reported use in either cardiovascular health or oncology. This is aligned with the ICTRP website that reports oncology and cardiovascular trials at the 1st and 3rd position for the numbers of trials by health category (the 2nd place being for neuropsychiatric conditions) [39]. We found half of drug trials (compared to other interventions) in our included studies with 12 publications of the 24 included reporting registry usage for drug trial pre-screening [44, 46,47,48, 51, 97]. One might argue that these benefits are not reflecting the current reality: since January 2014 there was a mean of 50,000 clinical trials running each year [39] and only 24 studies between 2014 and 2022 reported using population registries for pre-screening despite advantages with this method. However, the literature is known to under-report recruitment strategies in clinical trials, from protocols to publications [98, 99]. This leads to restrictive data, as this systematic review only reflects research that reported registry use in a clinical trial pre-screening setting. It is important to consider more clinical trials may pre-screen and recruit patients from registries without reporting it neither in their protocol nor in their published methodology. This means that registry use for trial pre-screening may be much more important than reported in this review. Furthermore, studies reporting use of population and disease registries for trial pre-screening have failed to address questions around data privacy and protection. The majority of disease registries around the world are accessible by two types of users: patients, who may directly fill out information into the registries, and health care professionals. These registries have data agreement in place, regarding privacy, sharing, and use such as data extraction for research purposes. When pre-screening for clinical trials, clinical trial sponsors do request pre-screening logs. This is done for financial reasons, as clinical research teams do negotiate in their clinical trial budget to be compensated for the time spent pre-screening patients for a specific trial. Pre-screening logs are provided by sponsors and collect limited data respecting information privacy regulations applying locally, such as General Data Protection Regulation (GDPR) in the European Union. It is essential to continue using tools such as pre-screening logs to serve as buffers to minimize data sharing from registries to sponsors (most often pharmaceutical companies) and maintain compliance with information privacy regulations. The main difference linked to this aspect would be observed between the USA and Europe, as the US regulation allows for race data to be collected which is not approved in Europe. This is limiting the evaluation of racial representativeness in European clinical trials, which may be biased by enrolling a vast majority of Caucasian participants.
Finally, as artificial intelligence (AI) is being developed, studies are now reporting use of machine learning for patient pre-screening into trials: Su et al. recently cited a pilot trial from the Mayo Clinic in Rochester using an AI-based trial matching system [2]. The paper reported an enrollment increase of 80% due to the quick and accurate patient matching to the oncology trial run at the Mayo Clinic [2], a system that could be applied to patient registries. Oncology has also brought us algorithms for clinical trial pre-screening, specifically Evolutionary Strategy algorithms (ES algorithms) [100, 101], that are commonly used in machine learning [102]. Ni et al. reported a 450% increase in efficacy of clinical trial pre-screening using electronic health record and not a patient registry, despite the fact that 10% of eligible patients were missed in the process [101]. More globally, data-driven technologies and strategies are more and more being reported in the literature, whether it is supporting prevention, diagnosis, or decision-making [103,104,105,106]. Such strategies’ impact on time optimization and associated cost reduction could be of great aid both to small trial centers working with limited staff and resources, and bigger trial centers dealing with a large volume of patients and trials.
Future studies are needed to address the limitations specific to certain disease fields to better describe the disease-specific needs around the use of registries for clinical trial pre-screening.
Conclusion
In conclusion, we aimed to describe the type of registries used, disease areas, type of clinical trials linked to the registry-based pre-screening, and potential assets the method brought to the pre-screening process. Only 24 studies between 2014 and 2022 reported using population and disease registries for clinical trial pre-screening despite time optimization and financial advantages using the method. A majority of the registries used were on a national level, and half of the trials for which pre-screening was performed were drug trials. Pre-screening strategies remain under-reported, and the use of population and disease registries for trial pre-screening may be much more important than what is described in this review, both for drug trials and non-drug trials. Our review is therefore stressing the need for standardized methodological guidelines for clinical trial pre-screening and encourages reporting of pre-screening processes in trial protocols and publications.
Availability of data and materials
Data is accessible upon request (full search and excel master document supporting screening, exported from Rayyan).
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- VC:
-
Vital capacity
- NCT:
-
National clinical trial
- SOPs:
-
Standard operating procedures
- EHR:
-
Electronic health record
- ICTRP:
-
International Clinical Trial Registry Platform
- WHO:
-
World Health Organization
- MeSH:
-
Medical subject heading
- LISA-R:
-
List of Included Studies and quality Assurance in Review
- RoB:
-
Risk of bias
- PECO:
-
Patient/Exposure/Comparator/Outcome
- PICO:
-
Patient/Intervention/Comparator/Outcome
- MND:
-
Motor neuron disease
- AI:
-
Artificial intelligence
- ES:
-
Evolutionary Strategy
References
Bramer WM, Giustini D, De Jonge GB, et al. De-duplication of database search results for systematic reviews in EndNote. jmla. 2016;104. https://doi.org/10.5195/jmla.2016.24.
Su Q, Cheng G, Huang J. A review of research on eligibility criteria for clinical trials. Clin Exp Med. 2023;23:1867–79.
Miesbach W, Oldenburg J, Klamroth R, et al. Gentherapie der Hämophilie: Empfehlung der Gesellschaft für Thrombose- und Hämostaseforschung (GTH). Hamostaseologie. 2023;43:196–207.
Superchi C, Brion Bouvier F, Gerardi C, et al. Study designs for clinical trials applied to personalised medicine: a sco** review. BMJ Open. 2022;12:e052926.
Chaytow H, Faller KME, Huang Y-T, et al. Spinal muscular atrophy: from approved therapies to future therapeutic targets for personalized medicine. Cell Rep Med. 2021;2:100346.
Sharma R. Innovative genoceuticals in human gene therapy solutions: challenges and safe clinical trials of orphan gene therapy products. CGT. 2024;24:46–72.
Tjeertes J, Bacino CA, Bichell TJ, et al. Enabling endpoint development for interventional clinical trials in individuals with Angelman syndrome: a prospective, longitudinal, observational clinical study (FREESIAS). J Neurodev Disord. 2023;15:22.
Sun Z, Zhang B, Peng Y. Development of novel treatments for amyotrophic lateral sclerosis. Metab Brain Dis. 2023. https://doi.org/10.1007/s11011-023-01334-z.
Gharat R, Dixit G, Khambete M, et al. Targets, trials and tribulations in Alzheimer therapeutics. Eur J Pharmacol. 2024;962:176230.
Revelles-Peñas L, Pastor-Navarro S, López-Piñero AA, Velasco-Tirado V. Use of a spinal cord stimulator to treat livedoid vasculopathy: Effective control of an untreatable disease. Estimulador medular en la vasculopatía livedoide: control eficaz de una patología intratable. Actas dermo-sifiliograficas. 2024:S0001-7310(23)00933-X. Advance online publication. https://doi.org/10.1016/j.ad.2023.02.037.
Goddard-Eckrich D, Gatanaga OS, Thomas BV, et al. Characteristics of drug-involved black women under community supervision; implications for retention in HIV clinical trials and healthcare. Soc Work Health Care. 2024;63:35–52.
Chaudhari N, Ravi R, Gogtay N, et al. Recruitment and retention of the participants in clinical trials: challenges and solutions. Perspect Clin Res. 2020;11:64.
Gul RB, Ali PA. Clinical trials: the challenge of recruitment and retention of participants. J Clin Nurs. 2010;19:227–33.
Hadidi N, Lindquist R, Treat-Jacobson D, et al. Participant withdrawal: challenges and practical solutions for recruitment and retention in clinical trials. Creat Nurs. 2013;19:37–41.
Juni P. Systematic reviews in health care: assessing the quality of controlled clinical trials. BMJ. 2001;323:42–6.
Hamel LM, Penner LA, Albrecht TL, et al. Barriers to clinical trial enrollment in racial and ethnic minority patients with cancer. Cancer Control. 2016;23:327–37.
Thoma A, Farrokhyar F, McKnight L, et al. Practical tips for surgical research: how to optimize patient recruitment. Can J Surg. 2010;53:205–10.
Pinto BM, Dunsiger SI. The many faces of recruitment in a randomized controlled trial. Contemp Clin Trials. 2021;102:106285.
on behalf of the INVESTIGATE studies group, Hilton P, Buckley BS, et al. Understanding variations in patient screening and recruitment in a multicentre pilot randomised controlled trial: a vignette-based study. Trials. 2016;17:522.
Parekh D, Patil VM, Nawale K, et al. Audit of screen failure in 15 randomised studies from a low and middle-income country. ecancer. 2022;16:1476. https://doi.org/10.3332/ecancer.2022.1476.
Mahajan P, Kulkarni A, Narayanswamy S, et al. Reasons why patients fail screening in Indian breast cancer trials. Perspect Clin Res. 2015;6:190.
Mckane A, Sima C, Ramanathan RK, et al. Determinants of patient screen failures in phase 1 clinical trials. Invest New Drugs. 2013;31:774–9.
Treweek S. Recruitment to trials - why is it hard and how might we make it less so? Trials. 2011;12:A110.
FDA. Enhancing the diversity of clinical trial populations — eligibility criteria, enrollment practices, and trial designs guidance for industry. Center for Biologics Evaluation and Research Center for Drug Evaluation and Research; 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/enhancing-diversity-clinical-trial-populations-eligibility-criteria-enrollment-practices-and-trial.
Wong SE, North SA, Sweeney CJ, et al. Screen failure rates in contemporary randomized clinical phase II/III therapeutic trials in genitourinary malignancies. Clin Genitourin Cancer. 2018;16:e233–42.
DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–85.
Lombardo G, Couvert C, Kose M, et al. Electronic health records (EHRs) in clinical research and platform trials: application of the innovative EHR-based methods developed by EU-PEARL. J Biomed Inform. 2023;148:104553.
Gilmore-Bykovskyi AL, ** Y, Gleason C, et al. Recruitment and retention of underrepresented populations in Alzheimer’s disease research: a systematic review. A&D Transl Res & Clin Interv. 2019;5:751–70.
Task Force Participants, Vellas B, Hampel H, et al. Alzheimer’s disease therapeutic trials: EU/US task force report on recruitment, retention, and methodology. J Nutr Health Aging. 2012;16:339–45.
Kirn DR, Grill JD, Aisen P, et al. Centralizing prescreening data collection to inform data-driven approaches to clinical trial recruitment. Alzheimers Res Ther. 2023;15:88.
Acuña-Villaorduña A, Baranda JC, Boehmer J, et al. Equitable access to clinical trials: how do we achieve it? Am Soc Clin Oncol Educ Book. 2023;43:e389838.
Gliklich RE, Dreyer NA, Leavy MB, editors. Registries for evaluating patient outcomes: a user’s guide. 3rd ed. Agency for Healthcare Research and Quality (US): Rockville; 2014. http://www.ncbi.nlm.nih.gov/books/NBK208616/. Accessed 16 Jan 2024.
Dreyer NA. Registries for robust evidence. JAMA. 2009;302:790.
Gabel RA. Patient registries, predictive models, and optimal care. JAMA. 2009;302:2662.
Bestehorn K. Medizinische Register: ein Beitrag zur Versorgungsforschung. Med Klin. 2005;100:722–8.
Williams WG. Uses and limitations of registry and academic databases. Semin Thorac Cardiovasc Surg Pediatr Card Surg Annu. 2010;13:66–70.
Williams WG, McCrindle BW. Practical experience with databases for congenital heart disease: a registry versus an academic database. Semin Thorac Cardiovasc Surg Pediatr Card Surg Annu. 2002;5:132–42.
Tan MH, Thomas M, MacEachern MP. Using registries to recruit subjects for clinical trials. Contemp Clin Trials. 2015;41:31–8.
Wold Health Organization W. International Clinical Trials Registry Platform (ICTRP). https://www.who.int/clinical-trials-registry-platform. Accessed 14 Dec 2023.
Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst Rev. 2021;10:89.
Clark JM, Sanders S, Carter M, et al. Improving the translation of search strategies using the Polyglot Search Translator: a randomized controlled trial. jmla. 2020;108. https://doi.org/10.5195/jmla.2020.834.
Lasch F, Weber K, Koch A. Commentary: on the levels of patient selection in registry-based randomized controlled trials. Trials. 2019;20:100.
ASCEND Study Collaborative Group, Aung T, Haynes R, et al. Cost-effective recruitment methods for a large randomised trial in people with diabetes: A Study of Cardiovascular Events iN Diabetes (ASCEND). Trials. 2016;17:286.
Achiron A, Givon U, Magalashvili D, et al. Effect of Alfacalcidol on multiple sclerosis-related fatigue: a randomized, double-blind placebo-controlled study. Mult Scler. 2015;21:767–75.
Heywood J, Evangelou M, Goymer D, et al. Effective recruitment of participants to a phase I study using the internet and publicity releases through charities and patient organisations: analysis of the adaptive study of IL-2 dose on regulatory T cells in type 1 diabetes (DILT1D). Trials. 2015;16:86.
Oni C, Mitchell S, James K, Ng WF, Griffiths B, Hindmarsh V, Price E, Pease CT, Emery P, Lanyon P, Jones A, Bombardieri M, Sutcliffe N, Pitzalis C, Hunter J, Gupta M, McLaren J, Cooper A, Regan M, Giles I, …. UK Primary Sjögren’s Syndrome Registry. Eligibility for clinical trials in primary Sjögren's syndrome: lessons from the UK Primary Sjögren's Syndrome Registry. Rheumatology (Oxford, England). 2016;55(3):544–52. https://doi.org/10.1093/rheumatology/kev373.
Darmon A, Bhatt DL, Elbez Y, et al. External applicability of the COMPASS trial: an analysis of the reduction of atherothrombosis for continued health (REACH) registry. Eur Heart J. 2018;39:750–757a.
Huebner H, Kurbacher CM, Kuesters G, et al. Heregulin (HRG) assessment for clinical trial eligibility testing in a molecular registry (PRAEGNANT) in Germany. BMC Cancer. 2020;20:1091.
van der Hout A, van Uden-Kraan CF, Holtmaat K, et al. Role of eHealth application Oncokompas in supporting self-management of symptoms and health-related quality of life in cancer survivors: a randomised, controlled trial. Lancet Oncol. 2020;21:80–94.
theFilnemus Myotonic Dystrophy Study Group, De Antonio M, Dogan C, et al. The DM-scope registry: a rare disease innovative framework bridging the gap between research and medical care. Orphanet J Rare Dis. 2019;14:122.
Heidrich B, Cordes H-J, Klinker H, et al. Treatment extension of pegylated interferon alpha and ribavirin does not improve SVR in patients with genotypes 2/3 without rapid virological response (OPTEX Trial): a prospective, randomized, two-arm, multicentre phase IV clinical trial. PLoS One. 2015;10:e0128069.
Toth GG, Lansky A, Baumbach A, et al. Validation of the all-comers design: results of the TARGET-AC substudy. Am Heart J. 2020;221:148–54.
Brown JC, Troxel AB, Ky B, et al. A randomized phase II dose–response exercise trial among colon cancer survivors: purpose, study design, methods, and recruitment results. Contemp Clin Trials. 2016;47:366–75.
Ashing K, Rosales M. A telephonic-based trial to reduce depressive symptoms among Latina breast cancer survivors: trial to reduce depressive symptoms. Psychooncology. 2014;23:507–15.
**ang JJ, Roy A, Summers C, et al. Brief report: implementation of a universal prescreening protocol to increase recruitment to lung cancer studies at a Veterans Affairs cancer center. JTO Clin Res Rep. 2022;3:100357.
Russo R, Coultas D, Ashmore J, et al. Chronic obstructive pulmonary disease self-management activation research trial (COPD–SMART): results of recruitment and baseline patient characteristics. Contemp Clin Trials. 2015;41:192–201.
Tamborlane WV, Chang P, Kollman C, et al. Eligibility for clinical trials is limited for youth with type 2 diabetes: insights from the Pediatric Diabetes Consortium T2D Clinic Registry. Pediatr Diabetes. 2018;19:1379–84.
Danila MI, Chen L, Ruderman EM, et al. Evaluation of an intervention to support patient-rheumatologist conversations about escalating treatment in patients with rheumatoid arthritis: a proof-of-principle study. ACR Open Rheumatol. 2022;4:279–87.
Green MF, Bell JL, Hubbard CB, McCall SJ, McKinney MS, Riedel JE, Menendez CS, Abbruzzese JL, Strickler JH, Datto MB. Implementation of a Molecular Tumor Registry to Support the Adoption of Precision Oncology Within an Academic Medical Center: The Duke University Experience. JCO Precis Oncol. 2021;5:PO.21.00030. https://doi.org/10.1200/PO.21.00030.
Guerra CE, Kelly S, Redlinger C, et al. Pancreatic cancer clinical treatment trials accrual: a closer look at participation rates. Am J Clin Oncol. 2021;44:227–31.
Wu J, Yakubov A, Abdul-Hay M, et al. Prescreening to increase therapeutic oncology trial enrollment at the largest public hospital in the United States. JCO Oncol Pract. 2022;18:e620–5.
Shadyab AH, LaCroix AZ, Feldman HH, et al. Recruitment of a multi-site randomized controlled trial of aerobic exercise for older adults with amnestic mild cognitive impairment: the EXERT trial. Alzheimers Dement. 2021;17:1808–17.
Mehta P, Raymond J, Han MK, et al. Recruitment of patients with amyotrophic lateral sclerosis for clinical trials and epidemiological studies: descriptive study of the National ALS Registry’s research notification mechanism. J Med Internet Res. 2021;23:e28021.
Valle CG, Camp LN, Diamond M, et al. Recruitment of young adult cancer survivors into a randomized controlled trial of an mHealth physical activity intervention. Trials. 2022;23:254.
Curtis JR, Wright NC, **e F, et al. Use of health plan combined with registry data to predict clinical trial recruitment. Clin Trials. 2014;11:96–101.
Laugesen K, Ludvigsson JF, Schmidt M, et al. Nordic Health Registry-based research: a review of health care systems and key registries. Clin Epidemiol. 2021;13:533–54.
Børø S, Thoresen S, Boge Brant S, et al. Initial investigation of using Norwegian health data for the purpose of external comparator arms - an example for non-small cell lung cancer. Acta Oncol. 2023;62:1642–8.
Axelsson L, Alvariza A, Lindberg J, et al. Unmet palliative care needs among patients with end-stage kidney disease: a national registry study about the last week of life. J Pain Symptom Manage. 2018;55:236–44.
Nørgaard M, Mailhac A, Fagerlund K, et al. Treatment patterns, survival, and healthcare utilisation and costs in patients with locally advanced and metastatic bladder cancer in Denmark 2015–2020. Acta Oncol. 2023;62:1784–90.
Bakken IJ, Ariansen AMS, Knudsen GP, et al. The Norwegian Patient Registry and the Norwegian Registry for Primary Health Care: research potential of two nationwide health-care registries. Scand J Public Health. 2020;48:49–55.
Pol T, Karlström P, Lund LH. Heart failure registries – future directions. J Cardiol. 2024;83:84–90.
Pålsson S, Pivodic A, Grönlund MA, et al. Cataract surgery in patients with uveitis: data from the Swedish National Cataract Register. Acta Ophthalmol. 2023;101:376–83.
Annual report 2021 with results and improvements measures from the national quality register for lung cancer. Oslo: The Cancer Registry of Norway; 2022. https://www.kreftregisteret.no/Generelt/Rapporter/Arsrapport-fra-kvalitetsregistrene/Arsrapport-for-lungekreft/.
Rimel BJ, Lester J, Sabacan L, et al. A novel clinical trial recruitment strategy for women’s cancer. Gynecol Oncol. 2015;138:445–8.
Kluding PM, Denton J, Jamison TR, et al. Frontiers: integration of a research participant registry with medical clinic registration and electronic health records. Clin Transl Sci. 2015;8:405–11.
Mudaranthakam DP, Thompson J, Hu J, et al. A Curated Cancer Clinical Outcomes Database (C3OD) for accelerating patient recruitment in cancer clinical trials. JAMIA Open. 2018;1:166–71.
Kannan V, Wilkinson KE, Varghese M, et al. Count me in: using a patient portal to minimize implicit bias in clinical research recruitment. J Am Med Inform Assoc. 2019;26:703–13.
McKinstry B, Sullivan FM, Vasishta S, et al. Cohort profile: the Scottish research register SHARE. A register of people interested in research participation linked to NHS data sets. BMJ Open. 2017;7:e013351.
Robotham D, Waterman S, Oduola S, et al. Facilitating mental health research for patients, clinicians and researchers: a mixed-method study. BMJ Open. 2016;6:e011127.
Vermunt L, Veal CD, Ter Meulen L, et al. European Prevention of Alzheimer’s Dementia Registry: recruitment and prescreening approach for a longitudinal cohort and prevention trials. Alzheimers Dement. 2018;14:837–42.
Zwan MD, Van Der Flier WM, Cleutjens S, et al. Dutch Brain Research Registry for study participant recruitment: design and first results. A&D Transl Res & Clin Interv. 2021;7:e12132.
Langbaum JB, Karlawish J, Roberts JS, et al. GeneMatch: a novel recruitment registry using at-home APOE genoty** to enhance referrals to Alzheimer’s prevention studies. Alzheimers Dement. 2019;15:515–24.
Grill JD, Hoang D, Gillen DL, et al. Constructing a local potential participant registry to improve Alzheimer’s disease clinical research recruitment. JAD. 2018;63:1055–63.
for the IMI-EPAD collaborators, Vermunt L, Muniz-Terrera G, et al. Prescreening for European Prevention of Alzheimer Dementia (EPAD) trial-ready cohort: impact of AD risk factors and recruitment settings. Alz Res Ther. 2020;12:8.
Harwood RH, Goldberg SE, Brand A, et al. Promoting Activity, Independence, and Stability in Early Dementia and mild cognitive impairment (PrAISED): randomised controlled trial. BMJ. 2023:e074787.
Jimenez-Maggiora GA, Bruschi S, Raman R, Langford O, Donohue M, Rafii MS, Sperling RA, Cummings JL, Aisen PS. TRC-PAD: Accelerating Recruitment of AD Clinical Trials through Innovative Information Technology. J Prev Alzheimers Dis. 2020;7(4):226–33. https://doi.org/10.14283/jpad.2020.48.
Langbaum JB, High N, Nichols J, Kettenhoven C, Reiman EM, Tariot PN. The Alzheimer's Prevention Registry: A Large Internet-Based Participant Recruitment Registry to Accelerate Referrals to Alzheimer's-Focused Studies. J Prev Alzheimers Dis. 2020;7(4):242–50. https://doi.org/10.14283/jpad.2020.31.
Milne R, Bunnik E, Tromp K, Bemelmans S, Badger S, Gove D, Maman M, Schermer M, Truyen L, Brayne C, Richard E. Ethical Issues in the Development of Readiness Cohorts in Alzheimer's Disease Research. J Prev Alzheimers Dis. 2017;4(2):125–31. https://doi.org/10.14283/jpad.2017.5.
Morgan RL, Whaley P, Thayer KA, et al. Identifying the PECO: a framework for formulating good questions to explore the association of environmental and other exposures with health outcomes. Environ Int. 2018;121:1027–31.
Kitchenham B. Guidelines for performing Systematic Literature Reviews in Software Engineering, Version 2.3, EBSE Technical Report EBSE-2007-01, Keele University and University of Durham. 2007.
Longinetti E, Regodón Wallin A, Samuelsson K, et al. The Swedish motor neuron disease quality registry. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19:528–37.
Treweek S, Pitkethly M, Cook J, et al. Strategies to improve recruitment to randomised trials. Cochrane Database Syst Rev. 2018;2018. https://doi.org/10.1002/14651858.MR000013.pub6.
Diguiseppi C, Goss C, Xu S, et al. Telephone screening for hazardous drinking among injured patients seen in acute care clinics: feasibility study. Alcohol Alcohol. 2006;41:438–45.
Graham A, Goss C, Xu S, et al. Effect of using different modes to administer the AUDIT-C on identification of hazardous drinking and acquiescence to trial participation among injured patients. Alcohol Alcohol. 2007;42:423–9.
Davis MK, Fine NM. An urgent need for data to drive decision making: rationale for the Canadian Registry for Amyloidosis Research. Can J Cardiol. 2020;36:447–9.
Ieva F, Gale CP, Sharples LD. Contemporary roles of registries in clinical cardiology: when do we need randomized trials? Expert Rev Cardiovasc Ther. 2014;12:1383–6.
Levesque E, Leclerc D, Puymirat J, et al. Develo** registries of volunteers: key principles to manage issues regarding personal information protection. J Med Ethics. 2010;36:712–4.
Lasagna L. Problems in publication of clinical trial methodology. Clin Pharmacol Ther. 1979;25:751–3.
O’Sullivan Greene E, Shiely F. Recording and reporting of recruitment strategies in trial protocols, registries, and publications was nonexistent. J Clin Epidemiol. 2022;152:248–56.
Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015;15:28.
Ni Y, Kennebeck S, Dexheimer JW, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc. 2015;22:166–78.
Kramer O. Evolution strategies. In: Machine learning for evolution strategies. Cham: Springer International Publishing; 2016. pp. 13–21. https://doi.org/10.1007/978-3-319-33383-0_2.
Rahman MA, Moayedikia A, Wiil UK. Editorial: data-driven technologies for future healthcare systems. Front Med Technol. 2023;5:1183687. https://doi.org/10.3389/fmedt.2023.1183687.
Subrahmanya SVG, Shetty DK, Patil V, Hameed BMZ, Paul R, Smriti K, Naik N, Somani BK. The role of data science in healthcare advancements: applications, benefits, and future prospects. Ir J Med Sci. 2022;191(4):1473–83. https://doi.org/10.1007/s11845-021-02730-z.
Sousa MJ, Pesqueira AM, Lemos C, Sousa M, Rocha Á. Decision-making based on big data analytics for people management in healthcare organizations. J Med Syst. 2019;43(9):290. https://doi.org/10.1007/s10916-019-1419-x.
Khan S, Khan HU, Nazir S. Systematic analysis of healthcare big data analytics for efficient care and disease diagnosing. Sci Rep. 2022;12(1):22377. https://doi.org/10.1038/s41598-022-26090-5.
Acknowledgements
Authors would like to thank the librarians from the Karolinska Institutet Library and especially Emma-Lotta Säätelä, for their help supporting the literature search for this study. We also thank Dr. Wim Grooten who acted as a mentor during this project in systematic review methodology.
Funding
Open access funding provided by Karolinska Institute. This study was funded by Stoppa ALS.
Author information
Authors and Affiliations
Contributions
JF and CI designed the study. JF and LA conducted the search and screened all citations. CI resolved screened conflicts when they arose. All authors participated to the manuscript, with JF and LA being major contributors to the manuscript writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
C. Ingre has consulted for Cytokinetics, Pfizer, BioArctic, Novartis, Tikomed, Ferrer, Amylyx, and Mitsubishi and was a DMC member for Appelis Pharmaceutical. She is also a board member of Tobii Dynavox and of the Stiching TRICALS Foundation; all outside the submitted work. All other authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Foucher, J., Azizi, L., Öijerstedt, L. et al. The usage of population and disease registries as pre-screening tools for clinical trials, a systematic review. Syst Rev 13, 111 (2024). https://doi.org/10.1186/s13643-024-02533-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13643-024-02533-0