Abstract
The light-harvesting protein complexes (Lhc) play key roles in the processes of light absorption and protection in diatoms. However, different Lhc protein carries out distinct function in photosynthesis. For now, roles of many Lhc proteins in light acclimation are largely unknown. Here, function of Lhcx3 in marine diatom Phaeodactylum tricornutum was examined by using reverse genetic technologies. The overexpression of Lhcx3 led to increased diadinoxanthin + diatoxanthin content and elevated non-photochemical fluorescence quenching (NPQ) while knockdown of Lhcx3 reduced NPQ level. In addition, the expression of Lhcx3 could be induced by blue light but not by red light. After addition of the photosynthetic inhibitor, upregulation of Lhcx3 transcript in high light could be inhibited by NH4Cl, but not by DCMU (3-(3,4-dichlorophenyl)-l,l-dim ethylurea). In contrast, DCMU addition increased expression of Lhcx3 in high light. In combination with changes of NPQ after addition of inhibitor, we concluded that the Lhcx3 played key roles in high light acclimation of diatoms. This finding will provide new clues for genetic improvement of P. tricornutum with an aim to cultivate new strains with high growth rate.
Similar content being viewed by others
Introduction
Diatoms are eukaryotic unicellular microalgae, and widely distributed in various marine environments (Field et al. 1998). They contribute greatly to the global primary production just like terrestrial tropical rain forests and grasslands, and serve as the base of the marine food webs (Armbrust 2009). Diatoms exhibit optimal photosynthetic activity over an extensive range of environments (Litchman et al. 2009). Their excellent adaptive capacity to changing conditions is regarded as a key reason why diatoms are predominant in contemporary oceans (Field et al. 1998). Levels of nonphotochemical quenching (NPQ) of diatoms under high light are much higher than those of higher plants (Roháček et al. 2014), showing their higher ability to dissipate excess energy (Ruban et al. 2004).
The key mechanism for the modulation of diatom photosynthesis is NPQ (Ruban and Murchie 2012). It occurs in the light-harvesting complex (Lhc) of PSII (Ruban and Murchie 2012). Under high light stress, excess light energy could be dissipated by some Lhc proteins as heat (Opačić et al. 2014). Lhc proteins in diatoms are also called fucoxanthin–chlorophyll proteins (Fcps). The Fcps can be divided into three groups: Lhcf, Lhcr, and Lhcx. Similar to the LhcSR proteins of Chlamydomonas reinhardtii, Lhcxs of diatoms have been shown to have the function of photoprotection (Bailleul et al. 2010; Dittami et al. 2010; Gundermann and Büchel 2012; Lepetit et al. 2013; Taddei et al. 2016). Using knockdown Phaeodactylum tricornutum, it is found that Lhcx1 plays a pivotal role in managing light responses (Bailleul et al. 2010). More recently, the roles of the other members of the Lhcx family in P. tricornutum were studied by using transgenic lines overexpressing the Lhcx proteins, revealing that different overexpressing lines with different Lhcx protein levels had a similar NPQ increase (Taddei et al. 2016). This probably suggests the complexity of NPQ regulation in diatoms. However, detailed roles for Lhcx2, Lhcx3 and Lhcx4 are still obscure.
At present, the genomes of P. tricornutum are publicly available (Bowler et al. 2008). P. tricornutum has been considered representative model species for studying response mechanism of diatoms to environmental changes, and also is one of the promising oleaginous algal species for biofuel production (Balamurugan et al. 2017; Wang et al. 2015). It is very important for lipid production of P. tricornutum to increase its biomass by regulating light-harvesting capacity. The Lhcx3 gene in P. tricornutum was found to be markedly upregulated in high light stress along with the increase of NPQ (Lepetit et al. 2013), suggesting that Lhcx3 took part in high light acclimation and photoprotection. But, to date, no evidence from reverse genetics directly supports the conclusion. Here, we showed that Lhcx3 played an important role in acclimation response of diatoms to high light by altering expression of Lhcx3. This finding will provide new clue for genetic improvement of P. tricornutum in efficiency of light energy utilization.
Materials and methods
Strain and culture conditions
Phaeodactylum tricornutum Bohlin CCMP-2561 was purchased from the Provasoli-Guillard National Center for Marine Algae and Microbiota, USA. P. tricornutum was incubated in natural seawater supplemented with f/2 (Guillard and Ryther 1962) without Na2SiO3∙9H2O. Microalgae were cultured in a climate incubator at 20 ± 0.5 °C under 12:12 h photoperiod with irradiance of 30 μmol m−2 s−1. High light (HL) treatments were implemented by irradiating the cells with 800 μmol m−2 s−1 for 6 h, then darkness treatments for 1 h. For experiments of light quality, cells were illuminated with monochromatic blue light (BL, 450 nm) or red light (RL, 520 nm). Low light (30 μmol m−2 s−1, LL), low medium light (100 μmol m−2 s−1, LML), and medium light (200 μmol m−2 s−1, ML) were used for BL or RL treatment. For experiments of inhibitor treatment, NH4Cl (5 mM, in water) and (3-(3,4-dichlorophenyl)-1,1-dimethylurea) DCMU (10 μM, in ethanol; final concentration of ethanol, 2‰) were added 20 min before starting the HL exposure. Cell density was counted by Brightline hemocytometer under an optical microscope. Cultures were collected at late log phase (day 7) for the following experiments.
Cloning, vector construction and algal transformation
The Lhcx3 mRNA sequence of P. tricornutum was retrieved from NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic analysis was performed by ClustalW2 and the phylogenetic tree was constructed by MEGA7.0 using neighbor-joining method. Total RNA from P. tricornutum was extracted using a plant RNA isolation kit (Omega, USA) and transcribed into cDNA using PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Japan) according to the manufacturer’s instruction. The full-length cDNA of Lhcx3 was PCR amplified using the Lhcx3-f/Lhcx3-r primer set. RNA knockdown vectors were constructed according to the method described by De Riso et al. (2009). Briefly, two fragments (short: 247 bp, corresponding to the PtLhcx3 gene sequence from 268 bp to 514 bp; long: 365 bp, corresponding to the PtLhcx3 gene sequence from 268 bp to 633 bp) of the full length PtLhcx3 cDNA were PCR amplified. All primers are provided in Additional file 1: Table S1.
Vector for Lhcx3 overexpression was generated by cloning the full-length cDNA sequence of the Lhcx3 gene into the Phy-21 vector (Siaut et al. 2007). The 633-bp fragment of Lhcx3 was purified and cloned into the diatom expression vector using the ClonExpress II one step kit (Vazyme, China), which yielded a recombinant expression vector Phy21-PtLhcx3. The Lhcx3 cDNA was inserted between the fucoxanthin chlorophyll a/c binding protein (fcp) fcpC promoter and fcpA terminator of P. tricornutum. Furthermore, in order to enhance transcriptional efficiency, an Omega leader sequence was placed before the coding region of PtLhcx3 (Fig. 1c). The recombinant vector was electroporated into P. tricornutum as previously described (Wang et al. 2015). Transformants were selected and maintained in f/2 medium supplemented with 30 μg mL−1 bleomycin (Ble).
Amino acid sequence analysis of PtLhcx3 and design of PtLhcx3 expression construct. a The conserved domain of PtLhcx3 amino acid sequence in P. tricornutum. The amino acid sequences of Lhcx1 (Phatr 27278) and Lhcx2 (Phatr 54065) from P. tricornutum, Lhcx2 (Thapsd_38879) and Lhcx LI818 (Thapsd 264921) from T. pseudonana, and LI818 (Chlre 184724) from Chlamydomonas reinhardtii were shown. b Phylogenetic tree showing evolutionary position of PtLhcx3. The top 25 hit were used as reference sequences when amino acid sequence of PtLhcx3 was searched against non-redundant protein database by BLASTP. Phylogenetic tree was constructed using the Neighbor-Joining method in MEGA7. The phylogenetic tree was tested by bootstrap** with 500 replicates and the percentages of bootstrap support were given on the branches. The PtLhcx3 was indicated by the blue arrow. c The schematic map showing cassettes of PtLhcx3 overexpression and knockdown driven by PfcpC promoter. An omega-leader was used for the enhancement of transcription
Detection of transformants
The Ble gene in transformants was detected by PCR using the Ble-f/Ble-r primer set (Additional file 1: Table S1). The relative abundance of PtLhcx3 transcript was quantified by quantitative real-time PCR (qRT-PCR) using SYBR Green qPCR SuperMix (Invitrogen, USA). Total RNA was isolated using Trizol (Invitrogen, USA), and reversely transcribed into first strand cDNA using Promega Reverse Transcription System (Promega, USA) with random primers according to the manufacturer’s instruction. The qPCR was performed in 8-strip PCR tubes on ABI PRISM® 7500 Sequence Detection System (ABI, USA). The relative gene expression was measured by the 2−ΔΔCt method after normalized to the internal reference gene, β-actin. The qPCR primers are provided in Additional file 1: Table S1.
For Western blot analysis, total protein was extracted using the Total Protein Extraction Kit (Vazyme, China). The protein content was quantitated by the BCA Protein Quantification Kit (Beyotime, China). Protein separation and electrotransfer were performed following standard procedures. Afterwards, the blot was incubated with primary anti-FLAG-tag antibody (1:3000, Abcam, USA) and HRP-conjugated goat anti-rabbit secondary antibody (1:5000, CST, USA), respectively. The membrane was developed using the chemiluminescent system. Endogenous β-actin was used as reference protein.
Chlorophyll fluorescence measurements
The chlorophyll fluorescence parameters of the transgenic and wild type (WT) cells were measured using PhytoPAM Phytoplankton Analyzer (WALZ, Germany) at room temperature. Effective quantum yield of PSII (ΦPSII) and relative electron transport rate (rETR) were read directly from PhytoPAM. NPQ was calculated using the formula: NPQ = (Fm – Fm′)/Fm′ (Bilger and Björkman 1990), where Fm and Fm′ are the maximum fluorescence emission levels in the dark and light-acclimated cells, respectively.
Detection of the reactive oxygen species (ROS) in cells
Intracellular ROS was detected using the dye dihydrorhodamine 123 (DHR123, Molecular Probes, Abcam) following a previous method (Jamers et al. 2009). Briefly, 100 μM DHR123 stock solutions were prepared in DMSO. And then, 10 μM DHR123 was freshly prepared in deionized water from stock solution before staining. Algal cells were incubated at room temperature for 20 min in the dark with DHR123 working solution. These stained cells were observed under a laser-scanning confocal microscope LSM880 (Zeiss, Germany) with an excitation wavelength of 507 nm, an emission wavelength of 529 nm.
Pigment analysis
For pigment analysis, 50 ml of culture was filtered on GF/F filter and the filter was immediately frozen in liquid nitrogen, and stored at − 80 °C until analysis. Pigment quantitation was performed using an Agilent 1200 HPLC system (Agilent technologies, USA) with a Symmetry C8 column as described previously (Jakob et al. 1999).
Statistical analysis
All experiments were performed at least in triplicate and the results were described as mean ± SD (standard deviation of the mean). Statistical comparison between transgenic lines and wild type was performed using Student’s t test. Difference with P < 0.05(*) or P < 0.01(**) was considered statistically significant.
Results
Sequence analysis of Lhcx3 in P. tricornutum
Amino acid sequences of the Lhcx3 (Phatr 44733) contained a conserved domain of chlorophyll a/b-binding which was predicted by InterPro (http://www.ebi.ac.uk) and indicated by blue frame in Fig. 1a. The signal peptide predicted by Signal P 4.1 was shown in red frame. Small difference of amino acid could be observed for the conserved domain of chlorophyll a/b-binding by aligning with other Lhcx proteins. Phylogenetic analysis showed that the PtLhcx3 (XP 002178699.1) had high similarity with the protein (XP 003079276.1) of Ostreococcus tauri (Fig. 1b). Furthermore, we predicted subcellular localization of the PtLhcx3 by using LocTree3, showing that the PtLhcx3 was localized to chloroplast membrane. In order to reveal function of Lhcx3, RNAi and overexpression vectors including promoter, 5′ leader, selectable marker were constructed (Fig. 1c).
Responses of PtLhcx3 to light quality and photosynthetic inhibitors
Under blue light (BL), the relative abundance of Lhcx3 transcript was significantly increased as light intensity increased, but did not change under red light (RL) (Fig. 2a). In addition, the ΦPSII in BL was lower than that in RL, but no significant difference was observed in rETR and NPQ between BL-treated and RL-treated cells when LL or LML was applied (Fig. 2b–d). However, under ML conditions, BL cultures had an increased capacity of NPQ and a larger rETR compared to RL cultures.
Responses of Lhcx3 (a), ΦPSII (b), rETR (c) and NPQ (d) of P. tricornutum to blue or red light. LL, LML, and ML represent the light intensities of 30 μmol m−2 s−1, 100 μmol m−2 s−1 and 200 μmol m−2 s−1, respectively. rETR is the relative electron translation rate. Significant difference is indicated at P < 0.05 (*) or P < 0.01 (**) level. Each value represents mean ± SD (n = 3)
After addition of the photosynthetic inhibitor, upregulation of Lhcx3 transcript under high light could be inhibited by NH4Cl, but not by DCMU (Fig. 3a). In contrast, DCMU addition led to an increased expression of Lhcx3 gene in high light. In addition, the increase of the rETR in high light (HL) was significantly inhibited by DCMU or NH4Cl addition (Fig. 3c). The reduction of the ΦPSII in HL was further strengthened by DCMU or NH4Cl addition (Fig. 3b). Nevertheless, the NPQ increase in HL was only prevented by NH4Cl, but not by DCMU.
Molecular characterization of transgenic lines
A 1-kb Ble gene fragment was PCR amplified from genomic DNA of transgenic cells, whereas no such fragment was detected in WT (Fig. 4a, b). This data showed that the expression cassette of the pHY21 has been successfully integrated into the host genome. qPCR analysis indicated that during HL, a significant increase and decrease were observed in relative abundance of Lhcx3 transcript in overexpressed (OE) and knocked-down (KNO) lines, respectively, compared to that of WT (Fig. 4c, d).
Molecular characteristics of transgenic lines. a Ble gene in overexpressed lines detected by genomic PCR; Lane M: 5000 bp marker, Lane 1, 2: A 1 kb band corresponding to Ble gene; Lane 3: genomic DNA of WT cells; Lane 4: water; b Ble gene in knocked-down lines detected by genomic PCR; Lane M: 5000 bp marker, Lane 1, 2: A 1 kb band corresponding to Ble gene; Lane 3: genomic DNA of WT cells; Lane 4: water; c Relative transcript level of Lhcx3 in overexpressed lines (OE1 and OE2) and wild type. Significant difference between WT and transgenic lines is indicated at P < 0.05 (*) or P < 0.01 (**) level. Each value represents mean ± SD (n = 3); d Relative transcript level of Lhcx3 in knocked-down lines (KNO1 and KNO2) and wild type. LL = 30 μmol m−2 s−1, HL = 800 μmol m−2 s−1
In addition, we detected expression of FLAG-tag ligated to the PtLhcx3 in overexpression vector in OE lines by using Western blot (Fig. 5). A specific cross-reactive band was observed in OE lines, whereas no band was detected in WT. We also observed that expression of FLAG-tag was increased gradually as light intensity went up from 30 to 800 μmol m−2 s−1 while no expression occurred in OE lines in the dark.
Phenotype changes of transgenic lines
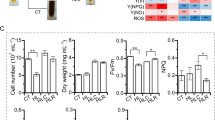
To analyze the effect of PtLhcx3 overexpression and knockdown on cellular physiological characteristics of transgenic lines, we determined the growth and chlorophyll fluorescence of the transgenic lines. Compared to WT cells, no significant difference was observed in growth rate, ΦPSII and rETR in PtLhcx3-overexpressed and knocked down lines in LL, HL or darkness (Figs. 6a–c and 7). However, during HL treatment, overexpression of PtLhcx3 led to an increased NPQ while knockdown of PtLhcx3 reduced NPQ (Fig. 6d).
To evaluate capacity to dissipate excess light energy in OE lines, we detected the level of ROS. ROS is induced by high light stress and its level can be measured using the ROS-sensitive dye, DHR123. ROS can oxidize DHR123 into the fluorescent derivative rhodamine 123. As shown in Fig. 8, obvious fluorescent signals were observed in WT cells, whereas they were hardly detected in OE lines.
The diadinoxanthin (Dd) and diatoxanthin (Dt) content was analyzed in WT and OE lines under LL or HL treatment (Table 1). Whether in LL or HL, both Dd and Dt content in OE lines was significantly higher than those in WT. However, after 1 h of HL exposure, the percentage of Dt was increased significantly in OE lines compared to WT while the percentage of Dd was reduced significantly.
Discussion
Diatoms have excellent capability to deal with changing light intensity in complicated marine environment. However, it is unclear how diatoms acclimate to drastic fluctuations in light intensity. For now, the Lhc proteins have been shown to play key roles in the processes of light absorption and protection in photosynthetic organisms (Govindjee 2002; Mozzo et al. 2008). Different Lhc proteins possess distinct functions. Thus, it is necessary to uncover roles of Lhc proteins in photosynthesis for providing new insights into processes of light energy utilization in diatoms.
Western-blot analysis of FLAG-tag indicated that Lhcx3 has been successfully expressed in OE lines. We also observed that LL, MHL or HL could induce expression of FlAG-tag, but band intensities were different, showing that the fcpC promoter was controlled by light. Considering that comparisons of physiological parameters between transgenic lines and WT were performed at the same light intensities, the difference of Lhcx3 expression under different light conditions did not affect their comparison. In transgenic lines with changed expression of PtLhcx3 under different light conditions, no significant changes were observed in the growth rate, ΦPSII and rETR compared to the WT, suggesting that PtLhcx3 might have nothing to do with cell growth and photosynthetic activity. In contrast, NPQ was significantly elevated or decreased in transgenic lines under HL, suggesting that PtLhcx3 may participate in photoprotection to prevent diatom cells from the damage of excess light. It has been reported that PtLhcx1 in the Lhcx family played a key role in the dissipation of excess light energy (Bailleul et al. 2010). But expression pattern of PtLhcx1 was different from that of PtLhcx3, it could not be induced by HL and was kept a high transcriptional level during the whole course of growth regardless of light conditions (Nymark et al. 2009). On the contrary, PtLhcx3 can be induced by HL. In photosynthesis, photosystems II (PSII) electrolyze H2O to produce proton gradient under HL and the LHC become aggregated due to the protonation of LHC and the de-epoxidation of diadinoxanthin to diatoxanthin (Goss et al. 2006). The aggregated LHC can dissipate excess light energy as heat to protect the photosynthetic apparatus from damage induced by HL (Walters et al. 2010). The induction of PtLhcx3 under HL might play a key role in triggering assembly of LHC.
The PtLhcx3 could be induced by BL, not by RL. Meanwhile, an increased NPQ was observed in BL cultures under ML conditions compared to RL cultures. A previous study found that P. tricornutum cultures grown under BL had an increased NPQ and a larger pool of xanthophyll cycle pigments compared to cultures grown under white light and RL (Costa et al. 2013), suggesting that BL induced an enhanced photoprotective potential. The upregulation of PtLhcx3 under BL conditions implicated that PtLhcx3 might be involved in photoprotection. Moreover, blue light receptor proteins such as CPF1 (Coesel et al. 2009) and CRYP (Juhas et al. 2014) have been found in P. tricornutum, and they may transmit light signal to regulate the expression of PtLhcx3.
After the addition of DCMU, the level of PtLhcx3 transcript in HL was significantly increased. DCMU is an inhibitor of electron transport chain of PSII, which can imitate a low light environment (Ridley and Horton 1984). When electron transfer was blocked by DCMU, electron accumulation will produce ROS in thylakoid (Lepetit et al. 2013). It is presumed that ROS induced the increase of PtLhcx3 transcript in HL. Notably, the addition of DCMU did not inhibit the increase of NPQ in HL, suggesting that PtLhcx3 might contribute to NPQ elevation. The reduced level of ROS in PtLhcx3 overexpression lines also supported the notion. The NH4Cl addition not only inhibited the level of PtLhcx3 transcript, but also reduced the increase of NPQ in HL. NH4Cl can remove the ΔpH of both sides of thylakoid membrane directly (Lovyagina and Semin 2016) while the induction of NPQ depends on the ΔpH (Eisenstadt et al. 2010). For now, it is still unclear how ΔpH induces NPQ. However, if sufficient PtLhcx3 protein is required for the development of NPQ, it is likely that changes of pH value in lumen of thylakoid affected the expression of PtLhcx3, which conversely influenced the formation of NPQ.
Compared to WT, both Dd and Dt content was raised significantly in OE lines regardless of light intensities. Meanwhile, after 1 h of HL exposure, Dt/(Dd + Dt) in OE lines was significantly higher than that in WT. These results suggested that increased expression of Lhcx3 in cells elicits de novo synthesis of the Dd + Dt pool. In T. pseudonana, it has been suggested that Lhcx6 was combined with Dt and participated directly in response of diatoms to HL (Zhu and Green 2010). Here, we hypothesized that the Dt in P. tricornutum may be combined with the Lhcx3. In this case, increased Lhcx3 required inevitably more Dt, which may provoke synthesis of more Dd for maintaining conversion between Dt and Dd. Given that Dt is vital for maintaining high level of NPQ during HL treatment (Eisenstadt et al. 2010), it is reasonable that an elevated NPQ occurred in Lhcx3-overexpressed lines. The results indicate there is a tight linkage among Lhcx3, Dt and NPQ.
References
Armbrust EV (2009) The life of diatoms in the world’s oceans. Nature 459:185–192
Bailleul B, Rogato A, De MA, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G (2010) An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci U S A 107:18214–18219
Balamurugan S, Wang X, Wang HL, An CJ, Li H, Li DW, Yang WD, Liu JS, Li HY (2017) Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol Biofuels 10:97
Bilger W, Björkman O (1990) Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res 25:173–185
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Todo T, Bowler C, Falciatore A (2009) Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep 10:655
Costa BS, Jungandreas A, Jakob T, Weisheit W, Mittag M, Wilhelm C (2013) Blue light is essential for high light acclimation and photoprotection in the diatom Phaeodactylum tricornutum. J Exp Bot 64:483–493
Dittami SM, Michel G, Collén J, Boyen C, Tonon T (2010) Chlorophyll-binding proteins revisited—a multigenic family of light-harvesting and stress proteins from a brown algal perspective. BMC Evol Biol 10:1–14
Eisenstadt D, Ohad I, Keren N, Kaplan A (2010) Changes in the photosynthetic reaction centre II in the diatom Phaeodactylum tricornutum result in non-photochemical fluorescence quenching. Environ Microbiol 10:1997–2007
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P (1998) Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240
Goss R, Pinto EA, Wilhelm C, Richter M (2006) The importance of a highly active and ΔpH-regulated diatoxanthin epoxidase for the regulation of the PS II antenna function in diadinoxanthin cycle containing algae. J Plant Physiol 163:1008–1021
Govindjee (2002) A role for a light-harvesting antenna complex of photosystem II in photoprotection. Plant Cell 14:1663–1668
Guillard RR, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8:229–239
Gundermann K, Büchel C (2012) Factors determining the fluorescence yield of fucoxanthin-chlorophyll complexes (FCP) involved in non-photochemical quenching in diatoms. BBA Bioenerg 1817:1044–1052
Jakob T, Goss R, Wilhelm C (1999) Activation of Diadinoxanthin De-Epoxidase Due to a Chiororespiratory Proton Gradient in the Dark in the Diatom Phaeodactylum tricornutum. Plant Biol 1:76–82
Jamers A, Lenjou M, Deraedt P, Bockstaele DV, Blust R, Coen Wd (2009) Flow cytometric analysis of the cadmium-exposed green alga Chlamydomonas reinhardtii (Chlorophyceae). Eur J Phycol 44:541–550
Juhas M, Von AZ, Spexard M, Schmidt M, Kottke T, Büchel C (2014) A novel cryptochrome in the diatom Phaeodactylum tricornutum influences the regulation of light-harvesting protein levels. FEBS J 281:2299–2311
Lepetit B, Sturm S, Rogato A, Gruber A, Sachse M, Falciatore A, Kroth PG, Lavaud J (2013) High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol 161:853–865
Litchman E, Klausmeier CA, Yoshiyama K (2009) Contrasting size evolution in marine and freshwater diatoms. Proc Natl Acad. Sci U S A 106:2665–2670
Lovyagina ER, Semin BK (2016) Mechanism of inhibition and decoupling of oxygen evolution from electron transfer in photosystem II by fluoride, ammonia and acetate. J Photochem Photobiol B 158:145–153
Mozzo M, Passarini F, Bassi R, Amerongen HV, Croce R (2008) Photoprotection in higher plants: the putative quenching site is conserved in all outer light-harvesting complexes of Photosystem II. BBA Bioenerg 1777:1263–1267
Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM (2009) An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4:e7743
Opačić M, Durand G, Bosco M, Polidori A, Popot JL (2014) Amphipols and photosynthetic light-harvesting pigment-protein complexes. J Membr Biol 247:1031–1041
Ridley SM, Horton P (1984) DCMU-induced fluorescence changes and photodestruction of pigments associated with an inhibition of photosystem I cyclic electron flow. Zeitschrift Für Naturforschung C 39:351–353
Riso VD, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A (2009) Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res 37:e96
Roháček K, Bertrand M, Moreau B, Jacquette B, Caplat C, Morantmanceau A, Schoefs B (2014) Relaxation of the non-photochemical chlorophyll fluorescence quenching in diatoms: kinetics, components and mechanisms. Philos Trans R Soc Lond B Biol Sci 369:20130241
Ruban AV, Murchie EH (2012) Assessing the photoprotective effectiveness of non-photochemical chlorophyll fluorescence quenching: a new approach. Biochim Biophys Acta 1817:977–982
Ruban A, Lavaud J, Rousseau B, Guglielmi G, Horton P, Etienne AL (2004) The super-excess energy dissipation in diatom algae: comparative analysis with higher plants. Photosynthesis Res 82:165
Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C (2007) Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406:23–35
Taddei L, Stella GR, Rogato A, Bailleul B, Fortunato AE, Annunziata R, Sanges R, Thaler M, Lepetit B, Lavaud J (2016) Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J Exp Bot 67:3939–3951
Walters RG, Ruban AV, Horton P (2010) Higher plant light-harvesting complexes LHCIIa and LHCIIc are bound by dicyclohexylcarbodiimide during inhibition of energy dissipation. FEBS J 226:1063–1069
Wang X, Liu YH, Hu DX, Balamurugan S, Lu Y, Yang WD, Liu JS, Li HY (2015) Identification of a putative patatin-like phospholipase domain-containing protein 3 (PNPLA3) ortholog involved in lipid metabolism in microalga Phaeodactylum tricornutum. Algal Res 12:274–279
Zhu SH, Green BR (2010) Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim Biophys Acta 1797:1449–1457
Authors’ contributions
HPD, LJO and TBH designed the study. TBH, TJ, XH and YFY carried out experiments and analyzed the data. HPD and TBH wrote the manuscript. HPD and LJO funded this study. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data analyzed during this study are included in this manuscript and additional material.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable. This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by the National Natural Science Foundation of China (41776121, 41576123), the Guangdong Natural Science Foundation (2018A030313164, 2016A030312004, 2015A030313326), and the International Science and Technology cooperation project (GASI-IPOVI-04), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA11020304).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
List of primers used in this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hao, TB., Jiang, T., Dong, HP. et al. Light-harvesting protein Lhcx3 is essential for high light acclimation of Phaeodactylum tricornutum. AMB Expr 8, 174 (2018). https://doi.org/10.1186/s13568-018-0703-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-018-0703-3