Abstract
Background
There is growing evidence that extracellular vesicles (EVs) play a crucial role in the paracrine mechanisms of transplanted human mesenchymal stem cells (hMSCs). Little is known, however, about the influence of microenvironmental stimuli on the osteogenic effects of EVs. This study aimed to investigate the properties and functions of EVs derived from undifferentiated hMSC (Naïve-EVs) and hMSC during the early stage of osteogenesis (Osteo-EVs). A further aim was to assess the osteoinductive potential of Osteo-EVs for bone regeneration in rat calvarial defects.
Methods
EVs from both groups were isolated using size-exclusion chromatography and characterized by size distribution, morphology, flow cytometry analysis and proteome profiling. The effects of EVs (10 µg/ml) on the proliferation, migration, and osteogenic differentiation of cultured hMSC were evaluated. Osteo-EVs (50 µg) or serum-free medium (SFM, control) were combined with collagen membrane scaffold (MEM) to repair critical-sized calvarial bone defects in male Lewis rats and the efficacy was assessed using µCT, histology and histomorphometry.
Results
Although Osteo- and Naïve-EVs have similar characteristics, proteomic analysis revealed an enrichment of bone-related proteins in Osteo-EVs. Both groups enhance cultured hMSC proliferation and migration, but Osteo-EVs demonstrate greater efficacy in promoting in vitro osteogenic differentiation, as evidenced by increased expression of osteogenesis-related genes, and higher calcium deposition. In rat calvarial defects, MEM with Osteo-EVs led to greater and more consistent bone regeneration than MEM loaded with SFM.
Conclusions
This study discloses differences in the protein profile and functional effects of EVs obtained from naïve hMSC and hMSC during the early stage of osteogenesis, using different methods. The significant protein profile and cellular function of EVs derived from hMSC during the early stage of osteogenesis were further verified by a calvarial bone defect model, emphasizing the importance of using differentiated MSC to produce EVs for bone therapeutics.
Similar content being viewed by others
Introduction
Bone tissue is a dynamic, intricate structure which undergoes continuous remodelling throughout an individual's lifetime. Disorders such as fractures and abnormalities can disrupt the normal bone-healing process, causing long-term morbidity and impairment. Current treatment approaches for bone healing include bone grafts and synthetic biomaterials. However, these techniques are not always effective and may lead to complications such as graft failure, infection, and immunological rejection [1]. There is increasing interest in the clinical potential of tissue engineering strategies using hMSC, biomaterials, and growth factors for bone regeneration [2]. While MSC have shown the ability to regenerate various tissues, the functional improvements observed after implantation do not always correlate with the number of MSC detected in situ [3]. This suggests that the secretome of MSC, which contains bioactive molecules such as cytokines, chemokines, growth factors and extracellular vesicles (EVs), such as microvesicles and exosomes, may modulate the effects of MSC on tissue regeneration through paracrine pathways [4].
The composition of MSC secretome or conditioned medium (MSC-CM) is highly dependent on the cell source and can be altered by various stimuli, such as hypoxia, cytokines, and serum deprivation. Several studies have shown that CM derived from MSC under different conditions can effectively promote bone healing and regeneration [5, 6]. EVs are among the key factors for these therapeutic effects. EVs are small vesicles (30–1000 nm in diameter) enclosed by a membrane, enriched with bioactive molecules such as lipids, proteins, and microRNAs [7]. While EVs isolated from MSC are reported to trigger bone healing [8], their therapeutic potential may vary according to the cellular origin, degree of differentiation and culture conditions. In recent years, there has been increasing interest in harnessing the therapeutic potential of EVs or exosomes to increase the osteogenic activity of MSC. Various studies have explored different strategies to achieve this, including exposing cultured MSC to osteogenesis-promoting factors or overexpressing osteogenic genes [8, 9]. Research has shown that exosomes released from MSC under osteogenic induction medium (OM) can upregulate genes related to osteogenesis and bone mineralization in vitro, indicating their potential as a therapeutic tool [9]. EVs derived from MSC treated with osteogenic media at different time points were found to be enriched with certain proteins and miRNAs with osteogenic properties, highlighting their potential as a source of therapeutic agents [10]. EVs derived from chemically or genetically osteogenically induced differentiated hMSC have been shown to induce the differentiation of undifferentiated MSC into osteoblasts in vitro [11]. This suggests that these EVs have remarkable osteogenic properties, which can be exploited for therapeutic purposes. Moreover, EVs released from hMSC during the late stage of osteogenic induction have shown significant potential in promoting osteogenic differentiation of undifferentiated MSC, further emphasizing their therapeutic potential [12]. However, isolating EVs or exosomes from the late stage of osteogenesis can be challenging, due to the disruption of their membrane structure caused by hydroxyapatite crystal formation [13]. Furthermore, EVs derived from mineralized primary osteoblasts have been observed to undergo changes in morphology and proteome profiles during in vitro osteogenesis, particularly in the late stage [14]. This highlights the importance of understanding the complex interplay between EVs and the osteogenic microenvironment. In this context, it is of interest to note that in a recent study, exosomes released from MSC during the early stage of osteogenesis promoted bone formation in mice with calvarial bone defects, highlighting the potential of early-stage EVs as a therapeutic tool [8].
A common clinical approach for treating advanced bone defects is guided bone regeneration (GBR), using occlusive barrier membranes, such as bioabsorbable collagen membranes (MEM) [15, 16]. These membranes function as a biocompatible barrier, which prevents the infiltration of competing soft tissue cells [17] and has the potential to serve as a carrier for various bioactive macromolecules, making them a promising delivery agent for bone formation applications [18]. We have recently reported the efficacy of MEM functionalized with hMSC-CM in rat calvarial bone defects, achieving bone formation comparable with that of implanted rat MSC [19]. It may be hypothesized that incorporating EVs into MEM can enable their sustained release and potentially improve the therapeutic efficacy of cell-free strategies for bone regeneration [20].
Despite the remarkable potential of EVs derived from undifferentiated or "naïve" MSC as an acellular therapy for bone defects, there remains a significant gap in knowledge regarding the properties and functions of EVs sourced from osteogenic differentiated MSC. In harvesting EVs, it is crucial to select carefully and expand a homogeneous population of cells with a known differentiation status: this is essential for obtaining reproducible results and producing EVs with predictable biological properties [14, 21]. Addressing these knowledge gaps is vital to achieving the full potential of acellular therapies using osteogenic EVs. The objectives of this study were therefore to investigate (a) the proteomic profile of EVs derived from naïve hMSC (Naïve-EVs) and MSC at an early stage of osteogenesis (Osteo-EVs), (b) the capacity of Osteo-EVs to promote in vitro hMSC proliferation, migration, and osteogenic differentiation compared with Naïve-EVs and (c) the capacity of Osteo-EVs to enhance in vivo bone formation in critical-sized rat calvarial defects. These investigations are intended to advance our knowledge of Osteo-EVs, with special reference to their potential as a novel acellular therapy for bone defects.
Materials and methods
Cell culture
The bone marrow mesenchymal stem (hMSC) used in this study was isolated from the anterior iliac crest of healthy human donors after ethical approval from the Regional Committee for Medical and Health Research Ethics in Norway (2013/1248/REK sør-øst C). The cells were expanded in growth medium (GM, Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 1% penicillin/streptomycin (GE Healthcare, South Logan, UT, USA) and 10% foetal bovine serum (FBS; GE Healthcare) at 37 °C in a humidified atmosphere, with 5% CO2. The medium was replaced twice a week. Based on the minimal criteria proposed by the International Society for Cellular Therapy (ISCT) to define hMSC, the cells were characterized for negative expression of surface marker antigens: CD34, HLA-DR, and CD45, and positive expression of surface marker antigens; CD73, CD90, and CD105, using flow cytometry. The cells were then tested for their multi-lineage differentiation potential into osteogenic, adipogenic, and chondrogenic lineages, using Alizarin Red S, Oil Red O, and Alican Blue staining methods, respectively, as described previously [22].
Preparation of osteogenic- and Naïve-conditioned media
To collect the conditioned medium (CM) from osteogenic and naïve (non-induced) culture conditions, at passages 3–5, hMSC from donors (n = 3) was trypsinized and seeded at 4000 cells/cm2 in GM. To generate CM from hMSC undergoing osteogenic differentiation, the cells were cultured in the osteogenic induction medium (OM) containing 100 nM dexamethasone, 45 μM ascorbic acid and 20 mM β-glycerophosphate (all from Sigma-Aldrich, St. Louis, MO, USA). The cells were maintained in this medium for 7 days. Next, the cells were washed three times with pre-warmed PBS and cultured in a serum-free medium (SFM), i.e. DMEM without supplements, for 3 days at 37 °C in 5% CO2, replaced with fresh SFM after 2 days. CM collected from the first 2 days and the third day was pooled and defined as an osteogenic-conditioned medium (Osteo-CM). To achieve a non-induced condition, after 7 days of incubation, the cells were cultured in SFM for 3 days at 37 °C in 5% CO2 and replaced after 2 days. After 3 days of incubation of cells in SFM, CM collected from the first 2 days and the third day was pooled and defined as a Naïve-conditioned medium (Naïve-CM). At the end of the experiment, collected CM was centrifuged at 4 °C (3000′ g for 5 min, followed by 2000′ g for 10 min) to remove cell debris and apoptotic bodies, followed by filtration through an 0.2-µm filter to remove larger particles. The prepared CM samples were then stored at − 80 °C.
Isolation of EVs
Size-exclusion chromatography (SEC) was used to isolate EVs from each group [23]. Briefly, Osteo- and Naïve-CM were concentrated using Amicon Ultra-15 Centrifugal Filter Units with Ultracel-100 membrane (MWCO = 100 kDa; Merck Millipore, USA) to ≤ 300 µl by repeated centrifugation at 3600 × g. The concentrate was adjusted to 500 µL using 0.2 µm-filtered and degassed PBS (dPBS), collected, and stored at − 80 °C as concentrated CM (CCM). Next, to isolate EVs from Naïve-CM (Naïve-EVs) and from Osteo-CM (Osteo-EVs), 500 µl of each respective CCM was applied to a 10-ml qEV column (35 nm qEVoriginal, no. 1000871, Izon Science Ltd) after columns had been equilibrated with 10 ml of PBS, as recommended by the manufacturer. The respective CCM was then pipetted onto the column to collect the output fractions. The first 3 ml (1st–6th fractions) was discarded as non-EV flowthrough, while the second 2.5 ml (7th–11th fractions) was collected as EV fractions and stored at – 80 °C until further analysis.
Characterization of Osteo- and Naïve-EVs
The immunophenotype of Osteo- and Naïve-EVs was analysed using immuno-affinity-based Dynabeads® magnetic separation technology via the flow cytometry method, to confirm the presence of EV-specific tetraspanin markers CD63, CD81, and CD9 (BD Bioscience, USA) according to the manufacturer’s recommendations (Life Technologies—Invitrogen, USA). Briefly, 10 µg of EVs from each group was resuspended in 1% BSA prepared in PBS and labelled with 20 µl of beads (Life Technologies—Invitrogen, USA) specific for each antibody (CD81, CD9, and CD63) by overnight incubation with gentle agitation. The solutions containing bead-bound EVs (EVs-beads) were then centrifuged and washed twice with 0.1% BSA. Finally, the EV beads were incubated with PE-labelled anti-human antibodies; CD63, CD81, and CD9, along with purified mouse IgG1, κ isotype control (BD Bioscience, USA) for 60 min at room temperature (RT). Data were acquired using an Accuri6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and analysed using FlowJo software (FlowJo V10.6.2). The size distribution of EVs was determined using dynamic light scattering (DLS, Zetasizer system, Malvern, UK). The protein concentration of EV samples was measured by Pierce™ BCA Protein assay kit (Thermo Fisher Scientific, USA), following the manufacturer’s instructions. EVs morphology was recorded by transmission electron microscopy (TEM), according to a previously described protocol [24]. The final EV preparations were examined using a Jeol JEM1400 transmission electron microscope (Jeol Ltd., Tokyo, Japan).
Liquid chromatography with tandem mass spectrometry (LC–MS/MS)
Sample preparation
Protein lysates collected from Osteo- and Naïve-EVs (n = 3 each) were analysed using LC–MS/MS with Label-Free Quantitation [25, 26]. In brief, approximately 25 μg of protein was digested into tryptic peptides. About 0.5 µg protein dissolved in 2% acetonitrile and 0.5% formic acid was injected into an Ultimate 3000 RSLC system connected online to an Exploris 480 mass spectrometer equipped with EASY-spray nano-electrospray ion source (all from Thermo Scientific, Sunnyvale, CA, USA) [26].
Bioinformatic analysis
LC–MS/MS raw data were searched using Proteome Discoverer software (version 435 2.5.0.400; Thermo Scientific). The Osteo- and Naïve-EVs protein datasets were then filtered and analysed for relative protein quantification among EV donors, using Perseus version 2.0.3.1 software. To ensure the precise quantification of proteins in each EV group, proteins that were not present in all three donors of each EV group were filtered out. The Venn diagram was used to compare the presence of common EV proteins in each EV group with the Extracellular vesicles database (including the top_100 proteins from Vesiclepedia data, http://microvesicles.org/) [27]. To identify the Gene Ontology Biological Process (GOBP), Gene Ontology Molecular Function (GOMF), and Gene Ontology Cellular Components (GOCC) associated with the common EV proteins of each EV group, a FunRich analysis tool (Version 3.1.3) was used. For statistical analysis, LIMMA was used (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4402510/) and DEqMS packages in R (v.4.2.1) (https://pubmed.ncbi.nlm.nih.gov/32205417/) to investigate differentially expressed proteins (DEPs) between Osteo- and Naïve-EVs. To predict GOBP, GOMF and Reactome pathways (REC) of DEPs in Osteo- and Naïve-EVs datasets, we used the Gene Ontology Resource version 17.0 (Panther) using Fisher's exact test as a type and false discovery rate (FDR) as a correction method [28]. To predict the potential participation of EV proteins in bone-related biological processes, we retrieved relevant bone biological process terms from QuickGO (https://www.ebi.ac.uk/QuickGO/, accessed in December 2022, EMBL-EMI, Cambridge, United Kingdom) and compared them with the names of DEPs and unique EV proteins (proteins not included in the statistical analysis) between Osteo- and Naïve-EVs.
In vitro osteogenic potential of EVs
Cell uptake
To determine the internalization of Osteo- and Naïve-EVs by the hMSC, 10 µg of Osteo- and Naïve-EVs, respectively, was labelled with a green fluorescent lipophilic dye- 3 mM DiO'; DiOC18 [3] (3,3′-Dioctadecyloxacarbocyanine Perchlorate/DMSO (DiOC18, Invitrogen™) for 2 h at 37 °C by gentle rotation [29]. Non-binding DiOC18 dye was then removed by diluting samples with PBS, followed by centrifugation using Vivaspin® 2, 100 kDa MWCO Polyethersulfone filters (Sartorius Stedim Biotech GmbH) at 300 × g for 3 min at 4 °C. hMSC was then washed twice with PBS and incubated at 37 °C with labelled EVs for 48 h, before fixing with 4% paraformaldehyde for 20 min. For staining of filamentous actin (F-actin), phalloidin–tetramethylrhodamine B isothiocyanate (Sigma-Aldrich, St. Louis, MO, USA) was added and incubated for 20 min at room temperature. Cellular nuclear DNA was then stained with 4′,6-diamidino-2-phenylindole (DAPI) for an additional 20 min. The cellular uptake of EVs was imaged using a Dragonfly 505 confocal spinning disc system (Andor Technologies, Inc, Belfast, Northern Ireland).
Cell proliferation
An Alamar blue cell viability assay (Thermo Fisher Scientific, Eugene, USA) was conducted after 24 and 72 h to assay cell proliferation. Briefly, hMSC was seeded at a density of 2 × 103 cells/100 μl of GM in a 96-well plate and cultured at 37 °C in 5% CO2 for 48 h. The hMSC was then washed with PBS and exposed to DMEM supplemented with 10% exosome-depleted FBS (FBS-ED-12F, Capricorn Scientific GmbH, Germany) and 10 μg/ml of Osteo- or Naïve-EVs, respectively, were added. All treatments were performed on day 1 (48 h after cell adhesion) and day 4 (72 h after the first dose). Next, 90 μl of fresh medium was mixed with 10 μl Alamar blue and incubated for 3 h at 37 °C with 5% CO2. The reduction of Alamar blue solution was measured by fluorescence values at 560 nm (excitation) and 590 nm (emission) using a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, Vantaa, Finland). Cell viability was assessed according to the following formula: Viability (%) = OD of experimental group/OD of control medium group × 100.
Cell migration
The effect of Osteo- and Naïve-EVs on MSC migration was evaluated by an in vitro wound-healing assay. Briefly, hMSC was seeded in a culture-insert (ibidi culture-insert 2 well, ibidi GmbH, Martinsried, Germany) at a density of 3 × 104 cells per well. When the cells reached over 100% confluence, the culture-insert was removed, and the cells were washed with PBS to remove non-adherent cells. To stop cell proliferation, 20 µg/ml of Actinomycin D-Ready Made Solution (Sigma-Aldrich, St. Louis, MO, US) was added and incubated at 37 °C in 5% CO2 for 2 h. The cells were washed with PBS and then different EV treatment groups were added. The hMSC was photographed at time points (t); t = 0 h, 24 and 48 h, in an inverted microscope (Eclipse TS100, Nikon, Tokyo, Japan). To track the wound closure, lines along the leading edges of each cell front were made, followed by measuring the decrease in the average distances of the lines using ImageJ software (version 1.52) [30]. The migration area was assessed using the following equation: migration area (%) = (A0 – Af)/A0 × 100, where A0 represents the initial wound area (t = 0 h) and Af represents the residual area of the wound at t = 24 h and 48 h, respectively [31].
Osteogenic differentiation
To study the effect of the different EVs on the osteogenesis of cultured hMSC, 10 µg/ml of Naïve- or Osteo-EVs was added to the OM. OM supplemented with an equal volume of PBS served as a control medium group. The media were changed twice a week, and EVs were added each time. After 14 days, alkaline phosphate staining (ALP) activity was tested using a BCIP/NBT alkaline phosphatase colour development kit, following the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, MUSA). After 21 days, calcium nodule formation was assessed by staining with 2% Alizarin Red Staining (ARS) (Sigma-Aldrich, St. Louis, MO, USA). Images were taken using a digital camera (ECLIPSE TS100, Nikon, Tokyo, Japan). To quantify calcium deposition, 10 mM% cetylpyridiniunm chloride (Sigma-Aldrich, St. Louis, MO, USA) was added and incubated for 20 min. Absorbance was then measured at 540 nm using a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, Vantaa, Finland). To determine the effect of the Osteo- and Naïve-EVs on the expression of osteogenesis-related genes in hMSC after 14 days of incubation, an osteogenic gene expression array and the validation of individual genes by qRT-PCR were conducted. Briefly, total RNA was extracted from hMSC using a Maxwell® 16 LEV simply RNA kit (Promega, Madison, WI, USA). RNA amount and purity were measured using Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The gene expression array (4,413,255, Applied Biosystems) and RT-qPCR, using TaqMan Fast Universal PCR Master Mix (Applied Biosystems), were assessed using a Step one™ Real-Time PCR System (Applied Biosystems). The array (4,413,255, Applied Biosystems, USA) was tailor-made (96 genes), covering, but not limited to, putative osteogenic differentiation markers, bone extracellular matrices, and TGF/BMP signalling pathways. The set of targeted genes and CT values are presented in Additional file 1. The expression levels were normalized by the following housekee** genes: RNA, 18S ribosomal 1 (18S rRNA), Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Hypoxanthine guanine phosphoribosyltransferase 1 (HPRT1) and beta-glucuronidase (GUSB). The expression of the osteogenesis-related human genes runt-related transcription factor 2 (RUNX2), collagen type I (Col 1A2), bone morphogenetic protein 2 (BMP-2), alkaline phosphatase (ALPL), osteopontin (SPP1), bone sialoprotein (BSP) and osteocalcin (BGLAP), was validated by real-time quantitative reverse transcription (RT-qPCR). The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as an endogenous control. The comparative Ct method was used for the relative measurement of gene expression level against the GAPDH gene. As shown in Additional file 2, all primers were from Applied Biosystems.
Effect of Osteo-EVs in vivo
Functionalization of MEM
To evaluate the effectiveness of MEM loaded with Osteo-EVs, on bone repair in vivo, a bilayer, non-cross-linked collagen membrane (25 mm × 25 mm; Bio‐Gide®, Geistlich Pharma, Wolhusen, Switzerland) was used. Briefly, MEMs were cut into small pieces (7 mm × 6 mm) using sterile scissors and placed with the “dense/smooth” surface facing the culture surface of the 48 -well plates and the “rough” surface facing upwards. Approximately 2.7 × 1010 particles of Osteo-EVs (50 µg in each 170 µl dPBS), or SFM as a control, were loaded onto each membrane and stored under sterile conditions at 4 °C for at least 4 h, to allow the Osteo-EVs to be absorbed [32]. The supernatants were then aspirated, and both Osteo-EVs- and SFM-loaded MEMs were stored at − 80 °C for lyophilization after adding a freeze–thaw protective agent mannitol (Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 0.5% (w/v). On the day of the animal experiments, lyophilized Osteo-EVs- and SFM-loaded MEMs were thawed and stored under sterile conditions at 4 °C until implantation.
Characterization of Osteo-EV-functionalized MEM
To evaluate the overall distribution of Osteo-EVs on MEM, Osteo-EVs- (50 ug/MEM) and SFM-loaded MEM were fixed with 2.5% glutaraldehyde in PBS for 30 min. Non-functionalized MEM served as controls. The fixed MEMs were washed three times with PBS and then dehydrated in an ascending series of ethanol. After evaporation of the ethanol, the samples were dried at room temperature, sputter-coated with gold–palladium and viewed in a scanning electron microscope (SEM) at 5 kV with a ZEISS SUPRA 55 VP. The average size of the Osteo-EVs in the MEM was determined using ImageJ 1.54 g.
Release of Osteo-EVs from MEM and uptake by cultured hMSC
The release of Osteo-EVs from MEMs and their uptake by hMSC were studied in vitro. Briefly, labelled Osteo-EVs/MEMs (10 µg) were positioned with the “rough” surface towards cultured hMSC in 8-ibidi wells (Nunc® Lab-Tek® Chamber Slide™ system). After 48 h, the Osteo-EVs/MEMs were removed and the cultured hMSC were washed twice with PBS, fixed with 4% paraformaldehyde for 15 min and stained with phalloidin–tetramethylrhodamine B isothiocyanate (Sigma-Aldrich, St. Louis, MO, USA) and DAPI as described above. The cells were then viewed in a Dragonfly 505 confocal spinning disc confocal scanning microscope (Andor Technologies, Inc, Belfast, Northern Ireland).
Calvarial defect model
All procedures were approved by the Norwegian Animal Research Authority (Mattilsynet; FOTS-17443) and conducted following the ARRIVE guidelines. Five male Lewis rats (LEW/OrlRj, Janvier Labs, Le Genest-Saint-Isle, France), 7 weeks old and weighing 200–250 g, were acclimated to standard vivarium conditions for two weeks. Before surgery, the animals were anaesthetized with a mixture of sevoflurane (Abbott Laboratories, Berkshire, UK) and O2 using a custom-made mask. As described previously [33], a 2-cm sagittal incision was made in the midline of the cranium and the periosteum was reflected to expose the parietal bones. Using a 5-mm outer-diameter trephine bur (Meisinger GmbH, Neuss, Germany), two full-thickness defects were created in each animal, one in each parietal bone. MEM loaded with Osteo-EVs (n = 5) or SFM (n = 5) as control was then randomly applied to the defects. The membranes were positioned with the “rough” surface towards the calvaria/defect (“dense/smooth” surface towards the soft tissue) and stabilized by applying ~ 5 μL of tissue adhesive (Histoacryl; B. Braun, Tuttlingen, Germany) at the corners. The rats were coded via ear clips following specific randomization. The allocation of treatments was adapted such that each animal received both treatments. After 2 weeks, an in vivo micro-computed tomography (µCT) scan was performed. At 4 weeks, the rats were euthanized with an overdose of CO2 and the calvaria was harvested and fixed in 4% paraformaldehyde. The primary outcome of interest was the new bone formation after 2 weeks using in vivo µCT and 4 weeks using ex vivo µCT and histology. For all experimental steps involving handling/analyses, animals and samples were identified by numbers to facilitate operators' blinding of treatment groups.
Micro-CT analysis
To follow the in vivo bone regeneration, the calvaria of the live rats was scanned 2 weeks after surgery by computed tomography (in vivo CT) under general anaesthesia, using a small-animal CT scanner and Mediso workstation (NanoScan Mediso, Budapest, Hungary): voxel size 40 μm, energy 70 kV, exposure 32 times 300 ms, projections 720, and 1:1 binning. For ex vivo μCT analysis, harvested samples were scanned using a SCANCO 50 μCT scanner (SCANCO Medical AG, Bruttisellen, Switzerland) at 90 kV and 200 μA, with an isotropic resolution of 20 μm. Both in vivo CT and ex vivo μCT scans were reconstructed using Amira software (Thermo Scientific) by orienting the drill direction along the Z-axis, with the defect in the approximate centre of the image. A standardized density threshold and volume of interest—including the entire thickness of the calvaria and excluding 0.5 mm of marginal bone—was defined for each defect. The percentage of new bone formation was calculated as a ratio of the total defect volume (BV/TV%). Bone coverage was calculated using ImageJ software (NIH) with custom-defined rulesets for CT and µCT scans.
Histology and histomorphometry
To visualize newly formed bone and soft tissues in the harvested samples 4 weeks after implantation, standardized thin-ground sections were prepared in the centre of each defect, parallel to the sagittal suture and perpendicular to the parietal bone. The samples were first dehydrated in an ascending series of alcohol and embedded in photocuring resin (Technovit 7200 + 1% benzoyl peroxide, Kulzer & Co., Wehrheim, Germany) before further processing using the EXAKT cutting and grinding equipment (EXAKT Apparatebau, Norderstedt, Germany). Undecalcified thin-ground sections were reduced to a thickness of approximately 100 µm and stained with Levi-Lazko dye (Morphisto GmbH, Frankfurt, Germany). The slices were scanned using an Olympus BX61VS digital virtual microscopy system (DotSlide 2.4, Olympus), with a 20 × magnification at a resolution of 0.32 μm per pixel. For histomorphometric analysis, respective areas of new bone without embedded MEM fibres (defined as new bone), new bone with embedded MEM fibres (defined as hybrid bone), total new bone (sum of new and hybrid bone), mineralized MEM fibres, residual MEM (non-mineralized MEM fibres), indefinite mineralized areas (defined as non-specific mineralization), which could not be attributed to any other category and soft tissue, were manually segmented and quantified in the central defect region using Adobe Photoshop (version 2022, Adobe, San Jose, CA, USA). Corresponding percentages of each tissue type were calculated as a ratio of the total central defect area. The central defect region was limited superiorly by the MEM, inferiorly by the dura mater and laterally by the defect edges.
Statistical analysis
All acquired data were tested by three donors and presented as the mean ± standard deviation. Multiple group comparisons were conducted by one-way ANOVA. Differences between the two groups were tested by Student’s t-test (normal distribution). Statistical analysis was undertaken using Graph Pad Prism 9.3.1 software (GraphPad, San Diego, CA, USA), and p < 0.05 was considered statistically significant.
Results
Isolation and characterization of hMSC and obtained EVs
hMSC was isolated and characterized based on immunophenotype and multi-differentiation capacity, according to the minimal criteria for defining hMSC, as proposed by ISCT [22, 34]. The isolated and collected Osteo- and Naïve-EVs were characterized. The average diameter of the Osteo-EVs particles (n = 3) was 162.8 nm (± 35.25 nm) (Fig. 1a i). The corresponding value for Naïve-EVs (n = 3) was 185 nm (± 41.76 nm) (Fig. 1b i). As observed by TEM, the Osteo- and Naïve-EVs displayed a typical cup-shaped vesicle (Fig. 1a, b, ii). Flow cytometry results confirmed the presence of EV surface markers CD63, CD81 and CD9 (> 95% each) (Fig. 1a, b, iii).
Characterization of Osteo- and Naïve-EVs. a i and b i Histogram graphs depicting the average size of EVs particles (n = 3) in Osteo- and Naïve-EVs, measured by dynamic light scattering (DLS). a ii and b ii Representative transmission electron microscopy (TEM) images of Osteo-and Naïve-EVs (Magnification of the EVs in the main image x10k; scale bar = 2 µm, and in the small box x70K; scale bar = 200 nm). a iii and b iii Representative flow cytometry analysis of transmembrane proteins CD63, CD81, and CD9. b i Venn diagram across all 3 donors in the Osteo-EVs group and inter-donor comparison, and across the common Osteo-EV (882) proteins and both Vesiclepedia Top 100 proteins and Vesiclepedia datasets. c ii The enriched GO terms (BP, CC, and MF) for the common Osteo-EVs (882) proteins using FunRich tool (Version 3.1.3). d i Venn diagram across all 3 donors in the Naïve-EVs group and compared to one another, and across the common Naïve-EV (895) proteins and both Vesiclepedia Top 100 proteins and Vesiclepedia datasets. d ii The enriched GO terms (BP, CC, and MF) for the common Naïve-EVs (895) proteins using FunRich tool (Version 3.1.3)
Proteomic analysis of Osteo- and Naïve-EVs
Horizontal bioinformatic analysis of the three donors of Osteo- and Naïve-EV groups
The proteomes of donor-matched Osteo- and Naïve-EVs from three independent donor-sample preparations were compared, using a label-free LC–MS/MS approach. The total number of proteins quantified for each of the two EV groups was similar: 1259 proteins in the Osteo-EVs and 1227 in the Naïve-EVs (Additional file 1). A Venn diagram was first used to analyse the common and different EV proteins of each EV group derived from three donors, D1, D2, and D3. In the Osteo-EVs group, 982, 1172 and 1070 proteins were identified in each respective donor: there were 882 common proteins, present in all three samples (Fig. 1c i). Among these 882 common proteins, 77 matched the typical Vesiclepedia top-100 markers, while 811 matched the whole EVs/exosome proteins in the Vesiclepedia database (Fig. 1c i). In the Naïve-EVs group, 1088, 1016, and 1099 proteins, respectively, were identified in each donor: there were 895 common proteins, present in all three samples (Fig. 1d i), of which 82 matched the typical Vesiclepedia top-100 markers and 829 matched the whole EVs/exosome proteins in the Vesiclepedia database, as shown in Fig. 1d i. Using the FunRich tool, Gene ontology enrichment analysis was used to analyse the common Osteo-EV (882) proteins and Naïve-EV (895) proteins involved in the GOBP, GOCC and GOMF terms (Fig. 1c, d, ii). The results showed that both common Osteo- and Naïve-EV proteins were related mainly to several GOCC, including “extracellular matrix, “centrosome”, “exosome”, “lysosomes”, and “cytoplasm”. Concerning GOMF, proteins of both EV groups were related to various similar terms, such as “Extracellular matrix structural constituent”, “Structural constituent of cytoskeleton”, “Chaperone and Ubiquitin-specific protease activities” and “Cytoskeletal protein binding”. For GOBP, the terms “Cell growth and/or maintenance”, “Protein metabolism”, “Energy pathways”, “Metabolism”, “Protein folding” and “protein metabolism,” were enriched in both EV groups. “Muscle development” and “Vesicle-mediated transport” were enriched only in the common Naïve-EVs group of proteins, while “Cell adhesion” and “Synapse organization and biogenesis” were enriched in the common Osteo-EV group of proteins.
Horizontal bioinformatics analysis of the Osteo- and Naïve-EV groups
A comparison of the proteins in the Osteo- and Naïve-EV groups (882 and 895, respectively) disclosed a total of 745 proteins common to both groups and 137 and 150 proteins, respectively, unique to the Osteo- and Naïve-EVs groups (Fig. 2a). These data are all presented in Additional file 1. The DEqMS method was then used for protein level differential analysis and to compare Osteo-EVs to Naïve-EVs (745 common proteins). This analysis resulted in the identification of a total of 187 DEPs: 96 upregulated in the Osteo-EVs group and 91 downregulated compared to Naïve-EVs (Volcano plot and Venn diagram, Fig. 2b i and ii). The data of all DEPs in each EV group are presented in Additional file 1. The proteome differences between the Osteo- and Naïve-EVs of all DEPs are visualized as a heatmap (Fig. 2c). To provide an overview of the GO annotation terms (BP, and MF) and the Reactome pathways of DEPs proteins among Osteo- and Naïve-EVs groups, the gene ontology resource Panther 17.0 was used for analysis [28]. The results showed that DEPs in the Osteo-EVs group were related mainly to several GOBP terms, of which “regulation of cell migration”, “regulation of cell motility and locomotion” and “developmental and cellular processes” were the most enriched processes (Fig. 2d i). In terms of GOMF, “binding and activity” functions were the most enriched terms. This includes “calcium ion binding, “protein binding”, “protein-containing complex binding”, and “growth factor binding” (Fig. 2d i). In terms of the REAC pathway, several pathways were enriched, including but not limited to “extracellular matrix organization”, “post-translational protein phosphorylation”, “platelet degranulation”, “regulation of Insulin-like Growth Factor (IGF) transport and uptake”, “elastic fibre formation”, “chondroitin sulfate/dermatan sulfate metabolism” and “Heparan Sulfate/Heparin (HS-GAG) metabolism” (Fig. 2d ii). In the Naïve-EVs group, in terms of GOBP, the DEPs were related mainly to “cellular process”, and to several related organization processes (e.g. “extracellular matrix and structure” and “external encapsulating structure”) (Fig. 2e i). In terms of GOMF, the DEPs were related to several function terms, of which those related to the bindings (e.g. “carbohydrate derivative”, “cell adhesion molecule” and “calcium ion”), and to “structural molecule activity” (Fig. 2e i). Several pathways related to the REAC pathway were identified, including the enriched pathways of "Extracellular matrix organization", "COPI-mediated anterograde transport", "extracellular matrix (ECM) proteoglycans", "homeostasis", as well as the pathways of "ER to Golgi Anterograde Transport" and "Transport to the Golgi and subsequent modification" (Fig. 2e ii).
Analysis of DEPs in proteins (745) common to Osteo- and Naïve-EVs groups. a Venn diagram representing the numbers of unique and overlap** proteins among the common protein in each EV group. b i and ii Volcano plot and Venn diagram showing differentially expressed proteins (p < 0.05) when comparing Osteo-EVs to Naïve-EVs. c Heat map showing the distribution of DEPs among the three donors of each EV group. Values represent protein level quantification (log2). d and e The enriched GO terms (BP, and MF) and REAC pathways for the DEPs in each EV group, respectively, are retrieved from the gene ontology resource
Osteo-EVs contain proteins involved in several osteogenesis-related biological processes
We next sought to identify the participation of the unique proteins and DEPs in each EV group in specific relevant and important biological processes involved in osteogenesis or bone healing. The gene names of the unique proteins and DEPs in the Osteo- and Naïve-EVs groups were compared with those of relevant biological processes from QuickGO annotation (https://www.ebi.ac.uk/QuickGO/). Table 1 lists the EV-identified proteins, including both unique proteins and the DEPs, which matched the selected biological processes. A total of 19 unique proteins and 8 DEPs identified in Osteo-EVs are associated with “cell population proliferation”, “cell migration”, “BMP signalling pathway”, “positive regulation of BMP signalling pathway”, and “Notch pathway”. Four unique proteins in Osteo-EVs are involved in “bone mineralization”, two unique proteins are associated with the “WNT signalling pathway” and most of the 19 unique proteins are associated with “cell migration”. However, none of the DEPs found in Osteo-EVs is associated with “bone mineralization” or the “WNT signalling pathway”. Moreover, Table 1 indicates that WNT5A and NOTCH2, two unique proteins found in Osteo-EVs, are involved in several selected biological pathways, according to QuickGO annotation. In contrast, the table demonstrates that all selected biological pathways are associated with the 12 unique proteins found in the Naïve-EVs group. Furthermore, only 5 DEPs in Naïve-EVs are associated with “cell population proliferation” and “cell migration” and none is associated with the “BMP signalling pathway”, “positive regulation of BMP signalling pathway”, “bone mineralization”, “Notch pathway”, or “WNT signalling pathway”. None of the DEPs found in Naïve-EVs was found to be linked to the “BMP signalling pathway”, “positive regulation of BMP signalling pathway”, “bone mineralization”, “Notch pathway” or the “WNT signalling pathway”.
Effect of Osteo- and Naïve-EVs on osteogenic differentiation of hMSC in vitro
Internalization of Osteo- and Naïve-EVs by hMSC and effect on their proliferation and migration in vitro
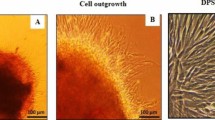
To exert their functional effects on target cells, EVs must come into contact with the cell membrane via specific receptors or be internalized by the cells. The data showed that both Osteo- and Naïve-EVs labelled with a green, fluorescent DiOC18 were internalized by hMSC after 48 h of culture compared with the control medium group (Fig. 3a). For cell proliferation, the results indicated that after 24 and 72 h, Osteo- and Naïve-EVs significantly stimulated hMSC proliferation compared with the control medium group, (*p < 0.05) (Fig. 3b). There were no statistically significant differences between the EVs groups. Similarly, for cell migration, the wound-healing assay results revealed a significant increase in cell migration in hMSC treated with Osteo- or Naïve-EVs after 48 h, compared to the control medium group (**p < 0.01, Fig. 3c i and ii). However, no significant differences were observed in cell migration between the hMSC groups treated with EVs, at either 24 or 48 h (Fig. 3c ii).
Effects of Osteo- and Naïve-EVs (10 µg/ml each) in cultured hMSC. a Representative images from the uptake experiments. Cultured hMSC internalised labelled Osteo- and Naïve-EVs: blue, DAPI-labelled nuclei; red, Phalloidin; green, DiOC18-labelled EVs. b Cell proliferation assay indicated that Osteo- and Naïve-EVs stimulate cultured hMSC proliferation after 24 and 72 h compared to the control medium group (BPS and medium). c i Representative images from the wound-healing experiments; scale bar 50 µm. Time course of cell migration wound-healing assay for cultured hMSC as the “recipient” cells with the control medium, Naïve- and Osteo-EVs. Scale bar 100 µm. c ii Quantification of the wound-healing assay from three independent donors. *p < 0.05, **p < 0.01
Effect of Osteo- and Naïve-EVs on osteogenic differentiation potential of hMSC
To compare the functional roles of Osteo- and Naïve-EVs in osteogenic differentiation, the hMSC was cultured in OM containing Osteo- or Naïve-EVs (10 µg/ml each), or an equal volume of PBS as a control, in a series of in vitro osteogenic differentiation-related assays. After 14 days, there was a slight increase in ALP in the hMSC treated with Naïve-EVs compared with the Osteo-EVs and control medium groups, as shown in Fig. 4a. After 21 days, ARS staining disclosed significantly greater formation of mineralized matrix in hMSC treated with Osteo-EVs than with Naïve-EVs and control medium (*p < 0.5) (Fig. 4b). To elucidate the genes underlying these effects, we examined the expression of mRNA encoding putative osteogenic differentiation markers, bone extracellular matrices, and molecules involved in TGF/BMP signaling pathways in hMSC, after 14 days of treatment, using a custom-made gene array (87 genes). Of these, 73 genes were detected in all samples and thus considered for further analysis, as shown on the heatmap (Fig. 4c). As shown in the prediction ellipses (Fig. 4d), analysis of gene expression across donors revealed that hMSC donors treated with Osteo-EVs exhibited the lowest variation, followed by the control medium and Naïve-EVs. In the hMSC treated with Osteo-EVs, four genes, namely MMP8, BMP2, IBSP, and MMP13, showed greater than twofold upregulation compared with the hMSC treated with control medium, whereas the Naïve-EVs group showed a trend towards downregulation of osteogenic gene expression (Fig. 4e). It is of interest to note that compared with the hMSC treated with Naïve-EVs, those treated with Osteo-EVs showed a trend towards upregulation of IBSP, MMP8, MMP13, BGLAP, SPP1, IGF1, COL4A3, COL4A4 and COL10A1, (Fig. 4f). In contrast, the hMSC treated with Naïve-EVs exhibited higher ALP expression than those treated with Osteo-EVs (Fig. 4f). RT-qPCR was used to verify the mRNA expression levels of RUNX2, Col1a2, ALP, BMP-2, SPP1, BSP, and BGLAP (Fig. 4g). The mRNA expression of RUNX2 was significantly downregulated in hMSC treated with EVs compared with the control medium (****p < 0.0001) and it was significantly downregulated in the hMSC treated with Osteo-EVs compared with those treated with Naïve-EVs (**p < 0.01). The mRNA expression level of Col1a2 was significantly downregulated only in the hMSC treated with Osteo-EVs, compared with the control medium (*p < 0.05). The mRNA expression of ALP was significantly higher in the hMSC treated with Naïve- than Osteo-EVs (*p < 0.05). Although the mRNA expression level of BMP2 was upregulated in the hMSC treated with Osteo-EV, the only significant level of expression was found in the hMSC treated with the Naïve-EVs compared with the control medium (*p < 0.05). The mRNA expression level of SPP1 was significantly higher in the hMSC treated with Osteo-EVs than in those treated with the control medium and Naïve-EVs (*p < 0.05, and **p < 0.01, respectively). The mRNA expression of BSP was significantly upregulated in the hMSC treated with Osteo-EVs compared with those treated with the Naïve-EVs (*p < 0.05). Similarly, the mRNA expression level of BGLAP was significantly upregulated in the hMSC treated with Osteo-EVs compared with those treated with control medium and Naïve-EVs (*p < 0.05) (Fig. 4g). Based on these in vitro findings, our next step was to conduct a preliminary investigation of the efficacy of Osteo-EVs in repairing bone defects in vivo.
Osteogenic effects of Osteo- and Naïve-EVs in cultured hMSC. a Representative images of ALP after 14 days. b Representative images of ARS after 21 days. Matrix mineralization was greater after Osteo-EVs treatment than after treatment with the Naïve-EVs and control medium. Quantification of the ARS experiment from three independent donors indicated that Osteo-EVs enhanced matrix mineralization of hMSC after 21 days. c Osteogenic gene expression array heatmap among the three donors of each group: control-medium, Naïve- and Osteo-EVs, after 14 days. d Prediction ellipses showed a new observation of donors from the same group falling within the prediction ellipse (confidence interval 95%). e Gene expression level (fold change) between the control medium and EV groups showed upregulation of several genes related to osteogenesis in the EV groups. Genes with red colours are identified in the Osteo-EVs and those with green colours are identified in the Naïve-EVs; green boxes refer to the downregulated genes in the Naïve-EVs f Gene expression level (relative fold change) between the Naïve- and Osteo-EVs showed upregulation of several genes related to osteogenesis in the Osteo-EVs group. g Validation of gene array findings with RT-qPCR: Osteo-EVs triggered higher mRNA expression levels of osteogenesis-related markers SPP1, BSP and BGLAP. *p < 0.05, **p < 0.01, ****p < 0.0001
In vivo osteogenic activity of Osteo-EVs
Adherence of Osteo-EVs to the collagen membrane and uptake by hMSC
SEM showed that the Osteo-EVs (2.7 × 1010 particles ≈ 50 µg/MEM) were well distributed over the MEM (Fig. 5a) with an average size of 61.47 nm (± 17.47 nm). To detect the release of Osteo-EVs from the MEM scaffolds and uptake by cultured hMSC, DiOC18 was used to label Osteo-EVs (10 µg/MEM). Dragonfly microscopy of the specimens, after 48 h of incubation, revealed the presence of DiOC18-labelled Osteo-EVs in the cytoplasm and perinuclear regions of hMSC (Fig. 5b), indicating that Osteo-EVs would be released from the MEM scaffold in vivo and then be taken up by resident cells.
Characterization of Osteo-EVs and MEMs in vitro and in vivo. a Representative SEM images of the MEM loaded with Osteo-EVs, compared with MEM loaded with only SFM. Scale bars, top: 2 µm, bottom: 200 nm with an average size of 61.47 nm (± 17.47 nm). b Representative images from the uptake experiments. Labelled Osteo-EVs were released from the MEM and entered targeted cultured hMSC after 48 h. MEM loaded with SFM served as a control. Blue, DAPI-labelled nuclei; red, Phalloidin; green, DiOC18-dye; scale bar 50 µm. c Representative reconstructed µCT images after 2 (in vivo) and 4 weeks (ex vivo) showing maximum, average, and minimum bone formation in Osteo-EVs and SFM groups. d Quantification of bone coverage (%) and bone volume per tissue volume (BV/TV %) in SFM and Osteo-EVs groups
Micro-CT analysis of new bone formation
The bone repair efficacy in vivo of Osteo-EVs loaded onto MEM was assessed in a critical-size rat calvarial bone defect model (Fig. 5c). All animals recovered from surgery and no adverse events were recorded. After 2 weeks, in vivo CT revealed greater coverage of the bone defects in Osteo-EVs (45.76 ± 22.17%) than SFM (30.69 ± 32.59%) (Fig. 5d). A similar trend was observed for BV/TV (1.34 ± 0.75% vs. 0.96 ± 1.03%) (Fig. 5d). At 4 weeks, bone formation was greater after implantation of Osteo-EVs than SFM (Fig. 5c). Although there were no significant differences, the relative coverage of the bone defects was 96.43 ± 3.67% for Osteo-EVs and 56.76 ± 40.50% for the SFM group (Fig. 5d). The level of BV/TV was 10.49 ± 3.06% in Osteo-EVs and 6.73 ± 6.53% in the SFM group, indicating that Osteo-EVs can improve ossification of defects.
Histological and histomorphometric analysis of new bone formation
After 4 weeks, both treatment groups, Osteo-EVs and SFM, exhibited a heterogeneous histological pattern combining the different tissue components after both treatments, as follows: new bone without incorporated MEM fibres, hybrid new bone with incorporated MEM collagen fibres, mineralized MEM fibres, residual MEM, soft tissues, and non-specific mineralization, which could not be attributed to hybrid bone or mineralized MEM fibres (Fig. 6a, b). At the base of the defect towards the dura, i.e. outside the MEM compartment, new bone was typically seen, characterized by well-structured woven bone (dark pink) or surrounded by layers of parallel-fibered bone (light pink) and an osteoid matrix (grey) (Fig. 6b). Adjacent to this newly formed bone an area of hybrid bone was observed, distinguished by immature woven bone, and incorporated collagen fibres from the MEM. Some degree of hybrid bone formation was also found next to the dura, more frequently in the Osteo-EVs/MEM (3/5) than in the SFM/MEM group (1/5). Frequently, hybrid bone was enclosed by new bone without MEM fibres, or in direct contact with residual MEM, indicating that hybrid bone was formed within the collagen MEM: ultimately this could be remodelled to the parallel-fibered bone. Residual MEM was evidently superior to the defect area, partially or completely covering the defect or as free-standing fibres integrated into the soft tissue among the bony networks. The histological evaluation further gave the impression that residual, unmineralized and mineralized fibres could be integrated into the hybrid-stage bone. All samples revealed some mineralized MEM fibres and/or non-specific mineralization. Finally, active bone formation was observed with characteristic osteoblast seams and osteoid (unstained; grey/white) in Osteo-EVs/MEM (5/5), compared with 3/5 in the SFM/MEM group. As shown in Fig. 6c, histomorphometric analysis revealed the presence of these different tissue types in all experimental groups, albeit in different proportions. Quantification of tissues in the central defect area revealed a greater new bone area in the Osteo-EVs/MEM (23.43 ± 7.02%) than in the SFM/MEM group (11.93 ± 9.00%; p = 0.054) (Fig. 6c). Quantification of total new bone (new bone + hybrid bone) revealed a non-significant trend (p = 0.052) in Osteo-EVs/MEM (46.95 ± 4.68%) compared with SFM/MEM (26.87 ± 25.07%; p = 0.110). hMSC treated with Osteo-EVs also exhibited lower areas of both mineralized MEM fibres (0.10 ± 0.57%) and residual MEM (9.70 ± 4.81%), compared with the SFM group, which exhibited mineralized MEM fibres (1.15 ± 1.54%) and residual MEM (24.75 ± 20.52%) (Fig. 6).
Micro-CT images, histological and histomorphometric analysis of MEM with Osteo-EVs. a Representative histological and μCT images of central slices at 4 weeks. Images showing maximum, average, and minimum bone formation in the Osteo-EVs and SFM groups, scale bar: 1 mm. b Representative histological images of Levi-Lazko dye staining at higher magnification, showing the different tissues analysed in the Osteo-EVs and SFM groups. The upper panel in each experimental group shows a region of interest with outlined sub-regions, which are enlarged in the lower panel (scale bars: upper 200 um, lower panel 50 um). Each sub-region shows a specific tissue type indicated by letters (A-F). (A and D) new bone (B and E), hybrid bone (C and F) and membrane. Numbers on the side panel indicate relative percentages of new bone for the treatment group (red), hybrid bone (cyan), mineralized membrane fibres (pink), residual unmineralized membrane (yellow) and soft tissue area (white). Yellow arrows; mineralized membrane, pink arrows; residual membrane. c Quantification of histomorphometric parameters. Data represent means (n = 5). SFM: serum-free medium; MEM: collagen membrane; Total bone (New Bone + Hybrid Bone Area): TtBAr; nB.Ar: New bone area; hB.Ar: Hybrid bone area; mMb.Ar: Mineralized membrane area; rMb.Ar: Residual Membrane area; Vd.Ar: Soft Tissue Area
Discussion
This study investigated extracellular vesicles, sourced from both osteogenic and naïve hMSC, with special reference to their potential to promote osteogenesis and bone formation [8]. While EVs from naïve hMSC show positive results, enhancing cell viability, proliferation, and bone formation, EVs derived from osteogenic differentiated hMSC show even more promising therapeutic effects. Nonetheless, it is important not to rely solely on the final outcome, but also to understand the characteristics and content profile of the EVs in question. Therefore, the main aim of this research was to improve understanding of the properties and functions of EVs derived from hMSC during the early stage of osteogenesis: the ultimate objective is to develop more effective acellular therapies for bone tissue disorders.
The current study shows similarities in morphology, size distribution, and expression of common transmembrane markers between Naïve- and Osteo-EVs, i.e. EVs obtained from hMSC during the early stage of osteogenic differentiation. In accordance with previous reports [35], these findings suggest that the brief period of osteogenic differentiation did not significantly alter the overall characteristics of the resultant EVs. MSC-EVs or exosomes exert their therapeutic effects in a content-dependent manner, with proteins, mRNAs and/or microRNAs playing a role in their function [36]. However, the primary focus of the present study was the protein signature of Osteo- and Naïve-EVs, because of their potential therapeutic effects on tissue repair/regeneration. It is important to note that in this study, no profiling of microRNAs was undertaken, despite their potential therapeutic effects [37], because of the limited amount of exosomal RNA available. Although the expression of EV proteins varied among donors, our analysis showed that most of the proteins were common to all. The isolated EVs were also identified by matching the proteins in both EV groups to the Vesiclepedia database. This suggests that the general characteristics of Osteo- and Naïve-EVs are similar and that the inherent cargo of MSC may play a significant role in determining the final EV products [38]. EVs derived from late-stage osteogenic differentiation of hBMSC also shared several proteins with EVs derived from naïve hBMSC [12]. Despite these similarities, we propose that the differentiation state of hMSC after 7 days, i.e. at an early stage of osteogenic differentiation, may result in EVs with different protein expressions. It is well documented that the contents of EVs are influenced by the cellular environment and their microenvironments, which in turn can alter the type and concentration of EV proteins and genetic materials such as microRNA [11, 37, "The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [67] partner repository, ProteomeCentral Datasets (proteomexchange.org), with the dataset identifier PXD043335". Supplementary data can be obtained by contacting the corresponding author. Human mesenchymal stem cells Undifferentiated hMSC-derived extracellular vesicles HMSC-derived extracellular vesicles during the early stage of osteogenesis Collagen membrane Mesenchymal stem cell-conditioned medium Osteogenic induction medium Serum-free medium Size-exclusion chromatography Liquid chromatography with tandem mass spectrometry Differentially expressed proteins Dioctadecyloxacarbocyanine Perchlorate 18 Wang W, Yeung KWK. Bone grafts and biomaterials substitutes for bone defect repair: a review. Bioact Mater. 2017;2(4):224–47. Gjerde C, Mustafa K, Hellem S, Rojewski M, Gjengedal H, Yassin MA, et al. Cell therapy induced regeneration of severely atrophied mandibular bone in a clinical trial. Stem Cell Res Ther. 2018;9(1):213. Iso Y, Spees JL, Serrano C, Bakondi B, Pochampally R, Song YH, et al. Multipotent human stromal cells improve cardiac function after myocardial infarction in mice without long-term engraftment. Biochem Biophys Res Commun. 2007;354(3):700–6. Konala VB, Mamidi MK, Bhonde R, Das AK, Pochampally R, Pal R. The current landscape of the mesenchymal stromal cell secretome: a new paradigm for cell-free regeneration. Cytotherapy. 2016;18(1):13–24. Al-Sharabi N, Xue Y, Udea M, Mustafa K, Fristad I. Influence of bone marrow stromal cell secreted molecules on pulpal and periodontal healing in replanted immature rat molars. Dent Traumatol Off Publ Int Assoc Dent Traumatol. 2016;32(3):231–9. Fujio M, **ng Z, Sharabi N, Xue Y, Yamamoto A, Hibi H, et al. Conditioned media from hypoxic-cultured human dental pulp cells promotes bone healing during distraction osteogenesis. J Tissue Eng Regen Med. 2017;11(7):2116–26. Fujita Y, Kadota T, Araya J, Ochiya T, Kuwano K. Clinical application of mesenchymal stem cell-derived extracellular vesicle-based therapeutics for inflammatory lung diseases. J Clin Med. 2018;7(10):355. Wang Y, Kong B, Chen X, Liu R, Zhao Y, Gu Z, Jiang Q. BMSC exosome-enriched acellular fish scale scaffolds promote bone regeneration. J Nanobiotechnol. 2022;20(1):444. Zhai M, Zhu Y, Yang M, Mao C. Human mesenchymal stem cell derived exosomes enhance cell-free bone regeneration by altering their miRNAs profiles. Adv Sci (Weinh). 2020;7(19):2001334. Pishavar E, Copus JS, Atala A, Lee SJ. Comparison study of stem cell-derived extracellular vesicles for enhanced osteogenic differentiation. Tissue Eng Part A. 2021;27(15–16):1044–54. Martins M, Ribeiro D, Martins A, Reis RL, Neves NM. Extracellular vesicles derived from osteogenically induced human bone marrow mesenchymal stem cells can modulate lineage commitment. Stem Cell Reports. 2016;6(3):284–91. Wang C, Stöckl S, Li S, Herrmann M, Lukas C, Reinders Y, et al. Effects of extracellular vesicles from osteogenic differentiated human BMSCs on osteogenic and adipogenic differentiation capacity of Naïve human BMSCs. Cells. 2022;11(16):2491. Ansari S, de Wildt BWM, Vis MAM, de Korte CE, Ito K, Hofmann S, Yuana Y. Matrix vesicles: role in bone mineralization and potential use as therapeutics. Pharmaceuticals (Basel). 2021;14(4):289. Davies OG, Cox SC, Azoidis I, McGuinness AJA, Cooke M, Heaney LM, et al. Osteoblast-derived vesicle protein content is temporally regulated during osteogenesis: implications for regenerative therapies. Front Bioeng Biotechnol. 2019;7:92. Sbricoli L, Guazzo R, Annunziata M, Gobbato L, Bressan E, Nastri L. Selection of collagen membranes for bone regeneration: a literature review. Materials (Basel). 2020;13(3):786. Dahlin C, Linde A, Gottlow J, Nyman S. Healing of bone defects by guided tissue regeneration. Plast Reconstr Surg. 1988;81(5):672–6. Sheikh Z, Sima C, Glogauer M. Bone replacement materials and techniques used for achieving vertical alveolar bone augmentation. Materials. 2015;8(6):2953–93. Ren Y, Fan L, Alkildani S, Liu L, Emmert S, Najman S, et al. Barrier membranes for guided bone regeneration (GBR): a focus on recent advances in collagen membranes. Int J Mol Sci. 2022;23(23):14987. Shanbhag S, Kampleitner C, Al-Sharabi N, Mohamed-Ahmed S, Apaza Alccayhuaman KA, Heimel P, et al. Functionalizing collagen membranes with MSC-conditioned media promotes guided bone regeneration in rat calvarial defects. Cells. 2023;12(5):767. Narayanan R, Huang CC, Ravindran S. Hijacking the cellular mail: exosome mediated differentiation of mesenchymal stem cells. Stem Cells Int. 2016;2016:3808674. Brown SV, Dewitt S, Clayton A, Waddington RJ. Identifying the efficacy of extracellular vesicles in osteogenic differentiation: an EV-lution in regenerative medicine. Front Dent Med. 2022;3:849724. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells: The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. Benedikter BJ, Bouwman FG, Vajen T, Heinzmann ACA, Grauls G, Mariman EC, et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci Rep. 2017;7(1):15297. Rana N, Suliman S, Al-Sharabi N, Mustafa K. Extracellular vesicles derived from primed mesenchymal stromal cells loaded on biphasic calcium phosphate biomaterial exhibit enhanced macrophage polarization. Cells. 2022;11(3):470. Aasebø E, Birkeland E, Selheim F, Berven F, Brenner AK, Bruserud Ø. The extracellular bone marrow microenvironment-a proteomic comparison of constitutive protein release by in vitro cultured osteoblasts and mesenchymal stem cells. Cancers (Basel). 2020;13(1):62. Al-Sharabi N, Gruber R, Sanz M, Mohamed-Ahmed S, Kristoffersen EK, Mustafa K, Shanbhag S. Proteomic Analysis of Mesenchymal Stromal Cells Secretome in Comparison to Leukocyte- and Platelet-Rich Fibrin. Int J Mol Sci. 2023;24(17). Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12): e1001450. Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–26. Nazarenko I, Rupp AK, Altevogt P. Exosomes as a potential tool for a specific delivery of functional molecules. Methods Mol Biol. 2013;1049:495–511. Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. Image J2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017;18(1):529. Masson-Meyers DS, Bumah VV, Enwemeka CS. Blue light does not impair wound healing in vitro. J Photochem Photobiol B. 2016;160:53–60. Zhang J, Liu X, Li H, Chen C, Hu B, Niu X, et al. Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway. Stem Cell Res Ther. 2016;7(1):136. Mohamed-Ahmed S, Yassin MA, Rashad A, Espedal H, Idris SB, Finne-Wistrand A, et al. Comparison of bone regenerative capacity of donor-matched human adipose-derived and bone marrow mesenchymal stem cells. Cell Tissue Res. 2021;383(3):1061–75. Mohamed-Ahmed S, Fristad I, Lie SA, Suliman S, Mustafa K, Vindenes H, Idris SB. Adipose-derived and bone marrow mesenchymal stem cells: a donor-matched comparison. Stem Cell Res Ther. 2018;9(1):168. Wang Z, He Z, Liang S, Yang Q, Cheng P, Chen A. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Res Therapy. 2020;11(1):511. Katsuda T, Ochiya T. Molecular signatures of mesenchymal stem cell-derived extracellular vesicle-mediated tissue repair. Stem Cell Res Ther. 2015;6:212. Wang X, Omar O, Vazirisani F, Thomsen P, Ekström K. Mesenchymal stem cell-derived exosomes have altered microRNA profiles and induce osteogenic differentiation depending on the stage of differentiation. PLoS ONE. 2018;13(2): e0193059. Aasebø E, Brenner AK, Hernandez-Valladares M, Birkeland E, Berven FS, Selheim F, Bruserud Ø. Proteomic comparison of bone marrow derived osteoblasts and mesenchymal stem cells. Int J Mol Sci. 2021;22(11):5665. Wei F, Li Z, Crawford R, **ao Y, Zhou Y. Immunoregulatory role of exosomes derived from differentiating mesenchymal stromal cells on inflammation and osteogenesis. J Tissue Eng Regen Med. 2019;13(11):1978–91. **n H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30(7):1556–64. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. Lai RC, Tan SS, Teh BJ, Sze SK, Arslan F, de Kleijn DP, et al. Proteolytic potential of the MSC exosome proteome: implications for an exosome-mediated delivery of therapeutic proteasome. Int J Proteomics. 2012;2012: 971907. Zhang B, Wang M, Gong A, Zhang X, Wu X, Zhu Y, et al. HucMSC-exosome mediated-Wnt4 signaling is required for cutaneous wound healing. Stem Cells. 2015;33(7):2158–68. Eirin A, Zhu XY, Puranik AS, Woollard JR, Tang H, Dasari S, et al. Comparative proteomic analysis of extracellular vesicles isolated from porcine adipose tissue-derived mesenchymal stem/stromal cells. Sci Rep. 2016;6:36120. Kim HS, Choi DY, Yun SJ, Choi SM, Kang JW, Jung JW, et al. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J Proteome Res. 2012;11(2):839–49. Wang C, Stöckl S, Li S, Herrmann M, Lukas C, Reinders Y, et al. Effects of extracellular vesicles from osteogenic differentiated human BMSCs on osteogenic and adipogenic differentiation capacity of naïve human BMSCs. Cells. 2022;11(16):2491. Chen J, Sun T, You Y, Wu B, Wang X, Wu J. Proteoglycans and glycosaminoglycans in stem cell homeostasis and bone tissue regeneration. Front Cell Dev Biol. 2021;9: 760532. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–28. Tao SC, Yuan T, Zhang YL, Yin WJ, Guo SC, Zhang CQ. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7(1):180–95. Salazar KD, Lankford SM, Brody AR. Mesenchymal stem cells produce Wnt isoforms and TGF-beta1 that mediate proliferation and procollagen expression by lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2009;297(5):L1002–11. Kumawat K, Menzen MH, Bos IS, Baarsma HA, Borger P, Roth M, et al. Noncanonical WNT-5A signaling regulates TGF-β-induced extracellular matrix production by airway smooth muscle cells. Faseb j. 2013;27(4):1631–43. Dong Y, Long T, Wang C, Mirando AJ, Chen J, O’Keefe RJ, Hilton MJ. NOTCH-mediated maintenance and expansion of human bone marrow stromal/stem cells: a technology designed for orthopedic regenerative medicine. Stem Cells Transl Med. 2014;3(12):1456–66. Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19(2):192–205. Itoh S, Udagawa N, Takahashi N, Yoshitake F, Narita H, Ebisu S, Ishihara K. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone. 2006;39(3):505–12. Shabbir A, Cox A, Rodriguez-Menocal L, Salgado M, Van Badiavas E. Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells Dev. 2015;24(14):1635–47. Zhu M, Liu Y, Qin H, Tong S, Sun Q, Wang T, et al. Osteogenically-induced exosomes stimulate osteogenesis of human adipose-derived stem cells. Cell Tissue Bank. 2021;22(1):77–91. Monaco E, Bionaz M, Hollister SJ, Wheeler MB. Strategies for regeneration of the bone using porcine adult adipose-derived mesenchymal stem cells. Theriogenology. 2011;75(8):1381–99. Ichida M, Yui Y, Yoshioka K, Tanaka T, Wakamatsu T, Yoshikawa H, Itoh K. Changes in cell migration of mesenchymal cells during osteogenic differentiation. FEBS Lett. 2011;585(24):4018–24. Al-Sharabi N, Xue Y, Fujio M, Ueda M, Gjerde C, Mustafa K, Fristad I. Bone marrow stromal cell paracrine factors direct osteo/odontogenic differentiation of dental pulp cells. Tissue Eng Part A. 2014;20(21–22):3063–72. Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF, et al. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int J Mol Sci. 2020;21(24):9739. Nakashima A, Tamura M. Regulation of matrix metalloproteinase-13 and tissue inhibitor of matrix metalloproteinase-1 gene expression by WNT3A and bone morphogenetic protein-2 in osteoblastic differentiation. Front Biosci. 2006;11:1667–78. Swanson WB, Zhang Z, **u K, Gong T, Eberle M, Wang Z, Ma PX. Scaffolds with controlled release of pro-mineralization exosomes to promote craniofacial bone healing without cell transplantation. Acta Biomater. 2020;118:215–32. Feher B, Apaza Alccayhuaman KA, Strauss FJ, Lee JS, Tangl S, Kuchler U, Gruber R. Osteoconductive properties of upside-down bilayer collagen membranes in rat calvarial defects. Int J Implant Dent. 2021;7(1):50. Caballé-Serrano J, Munar-Frau A, Delgado L, Pérez R, Hernández-Alfaro F. Physicochemical characterization of barrier membranes for bone regeneration. J Mech Behav Biomed Mater. 2019;97:13–20. Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177–83. Yan HC, Yu TT, Li J, Qiao YQ, Wang LC, Zhang T, et al. The delivery of extracellular vesicles loaded in biomaterial scaffolds for bone regeneration. Front Bioeng Biotechnol. 2020;8:1015. Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–52. "Mass spectrometry-based proteomic analyses were performed by the Proteomics Unit at the University of Bergen (PROBE). This facility is a member of the National Network of Advanced Proteomics Infrastructure (NAPI), which is funded by the Research Council of Norway (INFRASTRUKTUR-program project number: 295910)." Open access funding provided by University of Bergen. This work was supported by grants from the ITI (International Team for Implantology) Foundation, Switzerland (Grant No. 1417, 2019), and was partly supported by Trond Mohn Foundation, Norway (2021TMT08, 2022), and Meltzer Research Fund, Norway (13190000/ 103517129, 2022). “The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript”. NA-S conceived and designed the study. NA-S performed the experiments, data collection, and analysis, and drafted the manuscript. SM-A, SS, CK, RE, SY, NR, EB, ST, and RG assisted with experiments, sample preparation, data collection, data analysis, interpretation and/or drafting the manuscript. KM supervised the study, drafted the manuscript, and acquired funding. All authors read and approved the final version of the manuscript. Human cells and tissues were used within the framework of the research project of “Stem cells from bone marrow and adipose tissue for bone regeneration”, approved by the Regional Committees for Medical Research Ethics (REK) in Norway under reference number 2013-1248/REK sør-øst C, dated May 19, 2015. The use of animals was carried out under the approved project “Dental stem cells for regeneration of calvarial bone defects in rats” authorized by the Norwegian Animal Research Authority with reference number FOTS-17443 (Mattilsynet), date December 13, 2021. Not applicable. The authors declare no competing interests. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Mass spectrometry and gene expression array analysis data. . Overview of primers used for RT-qPCR. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data. Al-Sharabi, N., Mohamed-Ahmed, S., Shanbhag, S. et al. Osteogenic human MSC-derived extracellular vesicles regulate MSC activity and osteogenic differentiation and promote bone regeneration in a rat calvarial defect model.
Stem Cell Res Ther 15, 33 (2024). https://doi.org/10.1186/s13287-024-03639-x Received: Accepted: Published: DOI: https://doi.org/10.1186/s13287-024-03639-xAvailability of data and materials
Abbreviations
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Consent for publication
Competing interests
Additional information
Publisher's Note
Supplementary Information
Additional file 1.
Additional file 2: Table 1
Rights and permissions
About this article
Cite this article
Keywords