Abstract
Background
Although previous studies have suggested a close association between gut microbiota (GM) and intervertebral disc degeneration (IVDD), the causal relationship between them remains unclear. Hence, we thoroughly investigate their causal relationship by means of a two-sample Mendelian randomization (MR) study, aiming to determine the impact of gut microbiota on the risk of develo** intervertebral disc degeneration.
Methods
Summary data from genome-wide association studies of GM (the MiBioGen) and IVDD (the FinnGen biobank) have been acquired. The inverse variance weighted (IVW) method was utilized as the primary MR analysis approach. Weighted median, MR-Egger regression, weighted mode, and simple mode were used as supplements. The Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) and MR-Egger regression were performed to assess horizontal pleiotropy. Cochran's Q test evaluated heterogeneity. Leave-one-out sensitivity analysis was further conducted to determine the reliability of the causal relationship. A reverse MR analysis was conducted to assess potential reverse causation.
Results
We identified nine gut microbial taxa that were causally associated with IVDD (P < 0.05). Following the Benjamini–Hochberg corrected test, the association between the phylum Bacteroidetes and a higher risk of IVDD remained significant (IVW FDR-corrected P = 0.0365). The results of the Cochrane Q test did not indicate heterogeneity (P > 0.05). Additionally, both the MR-Egger intercept test and the MR-PRESSO global test revealed that our results were not influenced by horizontal pleiotropy (P > 0.05). Furthermore, the leave-one-out analysis substantiated the reliability of the causal relationship. In the reverse analysis, no evidence was found to suggest that IVDD has an impact on the gut microbiota.
Conclusion
Our results validate the potential causal impact of particular GM taxa on IVDD, thus providing fresh insights into the gut microbiota-mediated mechanism of IVDD and laying the groundwork for further research into targeted preventive measures.
Similar content being viewed by others
Introduction
Low back pain (LBP), which affects a staggering 70–85% of individuals at some point in their lives, presents a significant global public health challenge, resulting in a considerable financial burden on healthcare and social systems [1,2,3]. In general, LBP refers to discomfort, tension or inflexibility that is felt in the region of the body situated beneath the ribcage and above the inferior gluteal folds, often accompanied by leg pain (sciatica) and other neurological issues affecting the lower extremities [1, 4]. While various factors can play a role in the development of LBP, intervertebral disc degeneration (IVDD) stands out as one of the primary causes [4, 5]. IVDD serves as the pathological foundation for various spinal degenerative disorders and is a prevalent orthopedic condition that contributes to a reduced quality of life [6]. The intervertebral disc (IVD) consists of the nucleus pulposus (NP), annulus fibrosus, and the cartilage endplate, which are primarily composed of collagen and proteoglycan, imparting crucial properties to the disc. The NP is a critical component of the IVD, primarily made up of NP cells and the extracellular matrix (ECM). Intervertebral disc degeneration (IVDD) is a prevalent degenerative condition that is distinguished by the gradual reduction of proteoglycans and water content within NP [7]. As the disease progresses, the discs between the vertebrae may break down, rendering them more susceptible to herniation, which can cause compression of the spinal nerves and nerve roots. The irritation of nerves in the lower back as a result of IVDD is known as lumbar radiculopathy. If this occurs in the nerve roots of L4-S2, it commonly results in a distinct type of pain known as sciatica [8, 9].
The gut microbiota (GM) refers to the distinct microbial populations that inhabit the intestinal tract and coexist in a mutually beneficial relationship with the host organism, including bacteria, protozoa, fungi, archaea, and viruses [10]. It has the potential to influence multiple physiological processes, including metabolism, inflammation, and immune responses [11,12,13,14]. The identification of gut microbiota taxonomic characteristics and their potential role is mainly based on the utilization of 16S rRNA and metagenomic sequencing methods, which are commonly employed techniques [15]. In a recent study by Rajasekaran et al. [16], a total of 24 lumbar intervertebral discs (IVDs) were analyzed, revealing that the microbial makeup present in healthy IVDs contrasted with that of degenerated and herniated IVDs. Changes in the composition of the microbiome and the way hosts respond to microbiota, which can cause abnormal bone growth and resorption [17, 18], gave rise to the idea of the gut-bone marrow axis [19, 20] and the gut-bone axis [18]. Subsequent to the study conducted by Rajasekaran et al., a comparable gut-disc axis concept has emerged that could have significant implications in intervertebral disc degeneration and low back pain [16, 21]. As a result, the regulation of gut microbiota could potentially impact the diversity and quantity of microbiota within the intervertebral disc, ultimately hel** to regulate intervertebral disc degeneration.
However, additional investigation is required to further explore the distinct role of various gut microbiota taxa in the development of intervertebral disc degeneration. Akin to randomized controlled trials (RCT), the Mendelian randomization (MR) study is a recent research approach that investigates the causal relationship between exposure and outcome [22]. Mendelian randomization is a genetic epidemiology technique that uses single nucleotide polymorphisms (SNPs) that are known to affect modifiable exposures as instrumental variables (IVs) to deduce the causal effect of an exposure on an outcome. This approach is advantageous because it can eliminate confounding bias and can help to distinguish between the causal pathways of phenotypically grouped risk variables that are difficult to randomize or that are prone to measurement error [23]. In this study, we utilized GWAS summary statistics of GM and IVDD to perform MR analysis, with the aim of identifying GM taxa that may have a significant impact. This approach can help to confirm existing evidence and offer fresh perspectives on the management and prevention of intervertebral disc degeneration.
Materials and methods
Study design
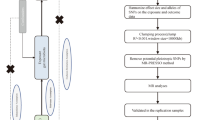
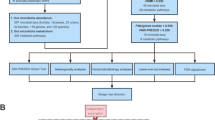
The overall flowchart of this study is shown in Fig. 1. MR studies require three assumptions to be met: (1) a strong correlation between the instrumental variable (IV) and the exposure, (2) IVs are unrelated to confounding factors, and (3) IVs are only related to the outcome through the exposure [24, 25]. Specifically, we determined the gut microbiota taxa that had a causal effect on intervertebral disc degeneration (IVDD) through bidirectional two-sample Mendelian randomization. Our results were reported according to the STROBE-MR guidelines [26]. We utilized GWAS data that had previously been obtained with informed consent and ethical approval for public release.
Data sources for exposure and outcome
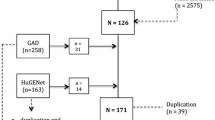
Based on twin, family, and population-based studies, it is evident that genetic factors also play a role in determining the composition of the gut microbiota, and some bacterial taxa exhibit heritability [27, 28]. The MiBioGen consortium conducted a study analyzing the genotypes of hosts and the sequencing profiles of 16S fecal microbiomes rRNA gene of 18,340 participants, as reported by Kurilshikov et al. [11]. The 18,340 participants in this study were sourced from 24 cohorts spanning the United States, Canada, Israel, South Korea, Germany, Denmark, the Netherlands, Belgium, Sweden, Finland, and the United Kingdom. Microbiome trait loci (mbTL) were identified using standardized methods to pinpoint genetic loci that influence the relative abundance (mbQTLs) or presence (mbBTLs) of specific microbial taxa. Finally, Gene Set Enrichment Analysis (GSEA) and Phenome-wide association studies (PheWAS) were conducted to provide biological interpretations of the results from the genome-wide association study (GWAS). The GWAS study examined 211 GM taxa ranging from genus to phylum level and discovered genetic variants associated with 9 phyla, 16 classes, 20 orders, 35 families, and 131 genera.
We obtained summary statistics of GWAS for IVDD from the FinnGen Consortium R8 release, which included 29,508 cases and 227,388 controls [29]. The FinnGen project was initiated in the autumn of 2017 in Finland, aiming to integrate genomic information with digital healthcare data and involving collaboration from universities, hospitals, THL (National Institute for Health and Welfare), blood service centers, biobanks, FINBB (Finnish Biobanks), international pharmaceutical companies, and hundreds of thousands of Finnish participants. The primary goal of this project is to collect and analyze genomic and health data from 500,000 participants in the Finnish Biobank to enhance human health and discover novel approaches for treating various diseases. The data collected includes individual clinical medical information, genomic data, and environmental/lifestyle factors. To establish an extensive and comprehensive research resource, FinnGen employs various data collection methods, including medical information from Finland’s national healthcare archives and genomic data obtained through large-scale gene sequencing and genoty** techniques. Additionally, the project may also gather disease-related survey questionnaires and biological specimens to obtain more comprehensive data information. The diagnosis of IVDD was based on ICD-10 M51, ICD-9 722, and ICD-8 275, excluded ICD-9 7220|7224|7227|7228A, ICD-8 7250. Table 1 presents detailed information on the exposure and outcome analyzed in this MR study. The details of the exposure and outcome are shown in Table 1.
Identification of IVs
SNPs closely associated with each GM taxon were used as instrumental variables (IVs) in this MR study. Due to the limited number of IVs obtained at a strict threshold (P < 5 × 10−8), a more comprehensive threshold (P < 1 × 10−5) was utilized to obtain a relatively higher number of IVs, thus resulting in more robust results [30]. In addition, to ensure the independence of each IV, SNPs within a 10,000 kb window size with a threshold of r2 < 0.001 were pruned to mitigate linkage disequilibrium (LD). Subsequently, we eliminated palindromic SNPs and SNPs that did not appear in the outcome from the IVs. Ultimately, we computed the F statistic for the IVs to evaluate the degree of bias due to weak instruments.The calculation formula is as follows: F = \(\frac{N - K - 1}{K} \times \frac{{R^{2} }}{{1 - { }R^{2} }}\); R2 = 2 × EAF × (1 − EAF) × BETA2 [31]. R2 represents the proportion of variance in the exposure that is explained by genetic variants. N = sample size; K = the number of IV. If the F statistic > 10, weak IVs were deemed not to have caused bias [32].
Statistical methods
For each GM taxon, the inverse variance weighted (IVW) method was used as the primary analysis method to determine causal associations (P < 0.05), with four additional methods (MR-Egger, weighted median, simple mode, and weighted mode) employed as supplementary measures [33, 34]. This study conducted sensitivity analyses in order to eliminate potential bias and examine the robustness of the IVW results. The Cochran Q test was utilized to assess heterogeneity among SNPs, and a P value greater than 0.05 indicates a lower likelihood of heterogeneity among the SNPs, in which case the IVW fixed-effect model was employed for analysis. Conversely, if the P value was less than or equal to 0.05, the IVW random-effects model was used [35].
In IVW regression, the intercept term is not considered and the reciprocal of the outcome variance (se2) is used as weights for fitting [33]. The weighted median method is defined as the median of the weighted empirical density function of the ratio estimates, and causal relationships can be consistently estimated if at least 50% of the information in the analysis comes from valid instruments [33]. In MR-Egger regression, the intercept term is considered, and the reciprocal of the outcome variance (se2) is also used as weights for fitting, with the resulting intercept used to assess horizontal pleiotropy [36]. MR-PRESSO global test was also utilized to achieve the same objective, which eliminated the influence of pleiotropy by removing outliers [37]. In addition, funnel plots and forest plots were constructed to visualize and ensure the reliability of the results. Finally, we converted the effect estimates to odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) to more intuitively display the causal associations between each GATA taxon and outcomes.
A significance level of P < 0.05 indicated the presence of a causal relationship between the exposure and outcome.To account for multiple testing (multiple exposures), the significance of the MR effect estimates was controlled using a Benjamini–Hochberg false discovery rate (FDR) of < 5% at a specific level. Additionally, we performed reverse causal analysis to examine the reverse causality relationship. To meet the core assumptions of MR, the selected SNPs were further filtered in the Phenoscanner database to ensure that the included instrumental variables were not correlated with known confounding factors [38], including dried fruit intake [57]. In the mouse model of IVDD, the abundance of Muribaculaceae and Lactobacillus increased, while the abundance of Clostridia_UCG-014 decreased. Furthermore, fecal microbiota transplantation further increased the abundance of Lactobacillus and reduced the abundance of Clostridia_UCG-014 [58]. Su et al. [59] conducted a Mendelian randomization study on the potential causal effects of specific gut microbiota and gut microbiota metabolites on low back pain (LBP). Similar to our findings, they observed that the abundance of the genus Marvinbryantia is a potential risk factor for LBP, while the abundance of the family Rikenellaceae and family Ruminococcaceae is a potential protective factor for LBP. These studies provide evidence for the association between gut microbiota and intervertebral disc degeneration (IVDD).
The pathogenic mechanisms underlying the role of gut microbiota in IVDD have been widely discussed in previous studies, involving various aspects such as inflammatory response, gut barrier function, and nutrient metabolism. These mechanisms intersect and collectively contribute to the overall impact of gut microbiota in IVDD. The Escherichia Shigella is a group of bacteria capable of producing lipopolysaccharide (LPS). LPS is a glycolipid component found in the outer membrane of gram-negative bacteria [60]. LPS can activate the TLR4/MyD88/NF-κB signaling pathway, leading to the release of pro-inflammatory mediators such as IL-6, IL-1β, and TNF-α. This cascade of events triggers a series of inflammatory processes, ultimately resulting in chronic low-grade inflammation [61]. A significant decrease in the abundance of a short chain fatty acids-producer, Marvinbryantia spp, was observed in the low muscle mass elders [62]. Sarcopenia is postulated to be an influential factor in chronic low back pain [87]. Further investigations are warranted to explore their potential involvement in IVDD. Finally, the association between human microbiota and the host in both healthy and disease states is a complex interplay rather than a simple one-way “causal relationship” [44]. Therefore, future studies should consider the intricate coordination and crosstalk between the host and gut microbiota to gain a better understanding of the relationship between gut microbiota and disease.
Conclusion
Using publicly available GWAS data, we conducted a bidirectional two-sample Mendelian randomization analysis on the causal association between 211 gut microbiota taxa and intervertebral disc degeneration (IVDD). Our analysis resulted in the identification of eight nominal causal associations and one strong correlation, further providing a theoretical foundation for the concept of the gut-disc axis. This study was based on a GWAS meta-analysis dataset generated from 16S rRNA sequencing, thus highlighting the need for analyses based on more advanced large-scale studies using metagenomics sequencing. Nevertheless, our research also offers valuable biomarkers for understanding the progression, diagnosis, and potential therapeutic approaches for IVDD.
Availability of data and materials
Please contact the corresponding author for data requests.
Abbreviations
- IVDD:
-
Intervertebral disc degeneration
- IVD:
-
Intervertebral disc
- NP:
-
Nucleus pulposus
- ECM:
-
Extracellular matrix
- GM:
-
Gut microbiota
- LBP:
-
Low back pain
- MR:
-
Mendelian randomization
- GWAS:
-
Genome-wide association studies
- IVW:
-
Inverse variance weighted
- WME:
-
Weighted median
- MR-PRESSO:
-
Mendelian randomization pleiotropy residual sum and outlier
- SNP:
-
Single nucleotide polymorphism
- IV:
-
Instrumental variable
- FDR:
-
False discovery rate
References
Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J, et al. What low back pain is and why we need to pay attention. Lancet (London, England). 2018;391(10137):2356–67.
Alonso-García M, Sarría-Santamera A. The economic and social burden of low back pain in Spain: a national assessment of the economic and social impact of low back pain in Spain. Spine. 2020;45(16):E1026-e1032.
Andersson GB. Epidemiological features of chronic low-back pain. Lancet (London, England). 1999;354(9178):581–5.
Knezevic NN, Candido KD, Vlaeyen JWS, Van Zundert J, Cohen SP. Low back pain. Lancet (London, England). 2021;398(10294):78–92.
Zehra U, Tryfonidou M, Iatridis JC, Illien-Jünger S, Mwale F, Samartzis D. Mechanisms and clinical implications of intervertebral disc calcification. Nat Rev Rheumatol. 2022;18(6):352–62.
Li Z, Chen X, Xu D, Li S, Chan MTV, Wu WKK. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52(6):e12704.
Raj PP. Intervertebral disc: anatomy-physiology-pathophysiology-treatment. Pain Pract. 2008;8(1):18–44.
Ropper AH, Zafonte RD. Sciatica. N Engl J Med. 2015;372(13):1240–8.
Jensen RK, Kongsted A, Kjaer P, Koes B. Diagnosis and treatment of sciatica. BMJ (Clin Res Ed). 2019;367:l6273.
Krga I, Glibetic M. 2.16—Gut microbiota in health and diseases. In: Glibetic M, editor. Comprehensive gut microbiota. Oxford: Elsevier; 2022. p. 182–98.
Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, Le Roy CI, Raygoza Garay JA, Finnicum CT, Liu X, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156–65.
Metwaly A, Reitmeier S, Haller D. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat Rev Gastroenterol Hepatol. 2022;19(6):383–97.
Cortes-Perez NG. 1.18—Microbiome and the immune system. In: Glibetic M, editor. Comprehensive gut microbiota. Oxford: Elsevier; 2022. p. 221–9.
Moszak M, Szulińska M, Bogdański P. You are what you eat—the relationship between diet, microbiota, and metabolic disorders—a review. Nutrients. 2020;12(4):1096.
Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021;11(1):3030.
Rajasekaran S, Soundararajan DCR, Tangavel C, Muthurajan R, Sri Vijay Anand KS, Matchado MS, Nayagam SM, Shetty AP, Kanna RM, Dharmalingam K. Human intervertebral discs harbour a unique microbiome and dysbiosis determines health and disease. Eur Spine J: Off Publ Eur Spine Soc Eur Spinal Deform Soc Eur Sect Cerv Spine Res Soc. 2020;29(7):1621–40.
Fernández-Murga ML, Olivares M, Sanz Y. Bifidobacterium pseudocatenulatum CECT 7765 reverses the adverse effects of diet-induced obesity through the gut-bone axis. Bone. 2020;141:115580.
Zaiss MM, Jones RM, Schett G, Pacifici R. The gut-bone axis: how bacterial metabolites bridge the distance. J Clin Investig. 2019;129(8):3018–28.
Santisteban MM, Kim S, Pepine CJ, Raizada MK. Brain-gut-bone marrow axis: implications for hypertension and related therapeutics. Circ Res. 2016;118(8):1327–36.
Vallianou NG, Geladari E, Kounatidis D. Microbiome and hypertension: Where are we now? J Cardiovasc Med (Hagerstown). 2020;21(2):83–8.
Li W, Lai K, Chopra N, Zheng Z, Das A, Diwan AD. Gut-disc axis: A cause of intervertebral disc degeneration and low back pain? Eur Spine J: Off Publ Eur Spine Soc Eur Spinal Deform Soc Eur Sect Cerv Spine Res Soc. 2022;31(4):917–25.
Swanson SA, Tiemeier H, Ikram MA, Hernán MA. Nature as a trialist? Deconstructing the analogy between mendelian randomization and randomized trials. Epidemiology. 2017;28(5):653–9.
Burgess S, Davey Smith G, Davies N, Dudbridge F, Gill D, Glymour M, Hartwig F, Holmes M, Minelli C, Relton C, et al. Guidelines for performing Mendelian randomization investigations [version 1; peer review: 1 approved, 1 approved with reservations]. 2019; 4(186).
Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–89.
Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods. 2019;10(4):486–96.
Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, VanderWeele TJ, Higgins JPT, Timpson NJ, Dimou N, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021;326(16):1614–21.
Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–43.
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in sha** human gut microbiota. Nature. 2018;555(7695):210–5.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. 2022.
Lv WQ, Lin X, Shen H, Liu HM, Qiu X, Li BY, Shen WD, Ge CL, Lv FY, Shen J, et al. Human gut microbiome impacts skeletal muscle mass via gut microbial synthesis of the short-chain fatty acid butyrate among healthy menopausal women. J Cachexia Sarcopenia Muscle. 2021;12(6):1860–70.
Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45(4):422–7.
Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–52.
Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28(1):30–42.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14.
Pagoni P, Dimou NL, Murphy N, Stergiakouli E. Using Mendelian randomisation to assess causality in observational studies. BMJ Ment Health. 2019;22(2):67–71.
Slob EAW, Groenen PJF, Thurik AR, Rietveld CA. A note on the use of Egger regression in Mendelian randomization studies. Int J Epidemiol. 2017;46(6):2094–7.
Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8.
Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations. Bioinformatics. 2019;35(22):4851–3.
Huang J, **e ZF. Dried fruit intake causally protects against low back pain: a Mendelian randomization study. Front Nutr. 2023;10:1027481.
** P, **ng Y, **ao B, Wei Y, Yan K, Zhao J, Tian W. Diabetes and intervertebral disc degeneration: a Mendelian randomization study. Front Endocrinol. 2023;14:1100874.
Shu P, Ji L, ** Z, Sun Z, Liu W. Association of insomnia and daytime sleepiness with low back pain: a bidirectional Mendelian randomization analysis. Front Genet. 2022;13:938334.
Zhou S, Zhu G, Xu Y, Gao R, Li H, Han G, Su W, Wang R. Mendelian randomization study on the putative causal effects of omega-3 fatty acids on low back pain. Front Nutr. 2022;9:819635.
Zhou J, Mi J, Peng Y, Han H, Liu Z. Causal associations of obesity with the intervertebral degeneration, low back pain, and sciatica: a two-sample Mendelian randomization study. Front Endocrinol. 2021;12:740200.
Kho ZY, Lal SK. The human gut microbiome—a potential controller of wellness and disease. Front Microbiol. 1835;2018:9.
Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. 2020;11:282.
De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2018;195(1):74–85.
Asquith M, Sternes PR, Costello ME, Karstens L, Diamond S, Martin TM, Li Z, Marshall MS, Spector TD, le Cao KA, et al. HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol (Hoboken, NJ). 2019;71(10):1642–50.
Boer CG, Radjabzadeh D, Medina-Gomez C, Garmaeva S, Schiphof D, Arp P, Koet T, Kurilshikov A, Fu J, Ikram MA, et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat Commun. 2019;10(1):4881.
Biver E, Berenbaum F, Valdes AM, Araujo de Carvalho I, Bindels LB, Brandi ML, Calder PC, Castronovo V, Cavalier E, Cherubini A, et al. Gut microbiota and osteoarthritis management: an expert consensus of the European society for clinical and economic aspects of osteoporosis, osteoarthritis and musculoskeletal diseases (ESCEO). Ageing Res Rev. 2019;55:100946.
Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, Furet J-P, Sokol H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76(9):1614–22.
Buchbinder R, van Tulder M, Öberg B, Costa LM, Woolf A, Schoene M, Croft P. Low back pain: a call for action. Lancet (London, England). 2018;391(10137):2384–8.
Feng Y, Egan B, Wang J. Genetic factors in intervertebral disc degeneration. Genes Dis. 2016;3(3):178–85.
Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, Abbasi-Kangevari M, Abbastabar H, Abd-Allah F, Abdelalim A, Abdollahi M. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet (London, England). 2020;396(10258):1204–22.
Hemanta D, Jiang XX, Feng ZZ, Chen ZX, Cao YW. Etiology for Degenerative Disc Disease. Chin Med Sci J = Chung-kuo i hsueh k'o hsueh tsa chih. 2016;31(3):185–91.
Khan AN, Jacobsen HE, Khan J, Filippi CG, Levine M, Lehman RA Jr, Riew KD, Lenke LG, Chahine NO. Inflammatory biomarkers of low back pain and disc degeneration: a review. Ann N Y Acad Sci. 2017;1410(1):68–84.
Kamali A, Ziadlou R, Lang G, Pfannkuche J, Cui S, Li Z, Richards RG, Alini M, Grad S. Small molecule-based treatment approaches for intervertebral disc degeneration: current options and future directions. Theranostics. 2021;11(1):27–47.
Fournier DE, Kiser PK, Shoemaker JK, Battié MC, Séguin CA. Vascularization of the human intervertebral disc: a sco** review. JOR spine. 2020;3(4):e1123.
Yao B, Cai Y, Wang W, Deng J, Zhao L, Han Z, Wan L. The effect of gut microbiota on the progression of intervertebral disc degeneration. Orthop Surg. 2023;15(3):858–67.
Su M, Tang Y, Kong W, Zhang S, Zhu T. Genetically supported causality between gut microbiota, gut metabolites and low back pain: a two-sample Mendelian randomization study. Front Microbiol. 2023;14:1157451.
Carnevale R, Pastori D, Nocella C, Cammisotto V, Bartimoccia S, Novo M, Del Ben M, Farcomeni A, Angelico F, Violi F. Gut-derived lipopolysaccharides increase post-prandial oxidative stress via Nox2 activation in patients with impaired fasting glucose tolerance: effect of extra-virgin olive oil. Eur J Nutr. 2019;58(2):843–51.
Mohammad S, Thiemermann C. Role of metabolic endotoxemia in systemic inflammation and potential interventions. Front Immunol. 2020;11:594150.
Han DS, Wu WK, Liu PY, Yang YT, Hsu HC, Kuo CH, Wu MS, Wang TG. Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin Nutr (Edinburgh, Scotland). 2022;41(7):1491–500.
Lin T, Dai M, Xu P, Sun L, Shu X, **a X, Zhao Y, Song Q, Guo D, Deng C, et al. Prevalence of sarcopenia in pain patients and correlation between the two conditions: a systematic review and meta-analysis. J Am Med Dir Assoc. 2022;23(5):902.e901-902.e920.
Ebenbichler G, Habenicht R, Ziegelbecker S, Kollmitzer J, Mair P, Kienbacher T. Age- and sex-specific effects in paravertebral surface electromyographic back extensor muscle fatigue in chronic low back pain. GeroScience. 2020;42(1):251–69.
Biddle A, Stewart L, Blanchard J, Leschine SJD. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5(3):627–40.
Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, Zhao J, Zhang H, Chen W. Blautia—a new functional genus with potential probiotic properties? Gut Microbes. 2021;13(1):1–21.
Hodgkinson K, El Abbar F, Dobranowski P, Manoogian J, Butcher J, Figeys D, Mack D, Stintzi A. Butyrate’s role in human health and the current progress towards its clinical application to treat gastrointestinal disease. Clin Nutr (Edinburgh, Scotland). 2023;42(2):61–75.
Liu H, Wang J, He T, Becker S, Zhang G, Li D, Ma X. Butyrate: A Double-Edged Sword for Health? Advances in nutrition (Bethesda, Md). 2018;9(1):21–9.
Chen M, Liao Z, Lu B, Wang M, Lin L, Zhang S, Li Y, Liu D, Liao Q, **e Z. Huang-Lian-Jie-Du-Decoction ameliorates hyperglycemia and insulin resistant in association with gut microbiota modulation. Front Microbiol. 2018;9:2380.
Ansari A, Bose S, Yadav MK, Wang JH, Song YK, Ko SG, Kim H. CST, an herbal formula, exerts anti-obesity effects through brain-gut-adipose tissue axis modulation in high-fat diet fed mice. Molecules (Basel, Switzerland). 2016;21(11):1522.
Zhang L, Wang Y, Wu F, Wang X, Feng Y, Wang Y. MDG, an Ophiopogon japonicus polysaccharide, inhibits non-alcoholic fatty liver disease by regulating the abundance of Akkermansia muciniphila. Int J Biol Macromol. 2022;196:23–34.
Wang Z, Wu H, Chen Y, Chen H, Wang X, Yuan W. Lactobacillus paracasei S16 alleviates lumbar disc herniation by modulating inflammation response and gut microbiota. Front Nutr. 2021;8:701644.
Kang C, Wang B, Kaliannan K, Wang X, Lang H, Hui S, Huang L, Zhang Y, Zhou M, Chen M, et al. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio. 2017;8(3):10–1128.
D’Amico D, Andreux PA, Valdés P, Singh A, Rinsch C, Auwerx J. Impact of the natural compound urolithin A on health, disease, and aging. Trends Mol Med. 2021;27(7):687–99.
D’Amico D, Olmer M, Fouassier AM, Valdés P, Andreux PA, Rinsch C, Lotz M. Urolithin A improves mitochondrial health, reduces cartilage degeneration, and alleviates pain in osteoarthritis. Aging Cell. 2022;21(8):e13662.
Liu H, Kang H, Song C, Lei Z, Li L, Guo J, Xu Y, Guan H, Fang Z, Li F. Urolithin A inhibits the catabolic effect of TNFα on nucleus pulposus cell and alleviates intervertebral disc degeneration in vivo. Front Pharmacol. 2018;9:1043.
Lin J, Zhuge J, Zheng X, Wu Y, Zhang Z, Xu T, Meftah Z, Xu H, Wu Y, Tian N, et al. Urolithin A-induced mitophagy suppresses apoptosis and attenuates intervertebral disc degeneration via the AMPK signaling pathway. Free Radical Biol Med. 2020;150:109–19.
Tabatabaeizadeh SA, Fazeli M, Meshkat Z, Khodashenas E, Esmaeili H, Mazloum S, Ferns GA, Abdizadeh MF, Ghayour-Mobarhan M. The effects of high doses of vitamin D on the composition of the gut microbiome of adolescent girls. Clin Nutr ESPEN. 2020;35:103–8.
Holick MF. High prevalence of vitamin D inadequacy and implications for health. Mayo Clin Proc. 2006;81(3):353–73.
Ramasamy B, Magne F, Tripathy SK, Venugopal G, Mukherjee D, Balamurugan R. Association of gut microbiome and vitamin D deficiency in knee osteoarthritis patients: a pilot study. Nutrients. 2021;13(4):1272.
Yang J, Li Y, Wen Z, Liu W, Meng L, Huang H. Oscillospira—a candidate for the next-generation probiotics. Gut Microbes. 2021;13(1):1987783.
Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology (Baltimore, MD). 2017;65(2):451–64.
Silwal P, Nguyen-Thai AM, Mohammad HA, Wang Y, Robbins PD, Lee JY, Vo NV. Cellular senescence in intervertebral disc aging and degeneration: molecular mechanisms and potential therapeutic opportunities. Biomolecules. 2023;13(4):686.
Ao X, Wang L, Shao Y, Chen X, Zhang J, Chu J, Jiang T, Zhang Z, Huang M. Development and characterization of a novel bipedal standing mouse model of intervertebral disc and facet joint degeneration. Clin Orthop Relat Res. 2019;477(6):1492–504.
Chen F, Liu H, Wang X, Li Z, Zhang J, Pei Y, Zheng Z, Wang J. Melatonin activates autophagy via the NF-κB signaling pathway to prevent extracellular matrix degeneration in intervertebral disc. Osteoarthr Cartil. 2020;28(8):1121–32.
Zhang GZ, Deng YJ, **e QQ, Ren EH, Ma ZJ, He XG, Gao YC, Kang XW. Sirtuins and intervertebral disc degeneration: roles in inflammation, oxidative stress, and mitochondrial function. Clin Chim Acta; Int J Clin Chem. 2020;508:33–42.
Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021;19(8):514–27.
Acknowledgements
Not applicable.
Funding
The study is supported by the National Natural Science Foundation of China (81472140), the Natural Science Foundation of Tian** City (S20ZDD484) and Health and Technology Project of Tian** Municipal Commission, Grant No. zc20219.
Author information
Authors and Affiliations
Contributions
ZMG and JM contributed to conception and design of the study. ZMG, JW and GDC organized the database. ZMG and JW performed the statistical analysis. ZG wrote the first draft of the manuscript. JCL, JL and ZPZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version. All author(s) read and approved the final manuscript. Jun Miao conceived and designed the study.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We utilized GWAS data that had previously been obtained with informed consent and ethical approval for public release.
Consent for publication
Written informed consent for publication was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Table S1. Instrument Variables for all gut microbiota taxa. Table S2. Casual effects of MR analysis between gut microbiota and intervertebral disc degeneration. Table S3. Reverse MR analysis for the causal effect of intervertebral disc degeneration on gut microbiota.
Additional file 2.
Fig. S1. Leave-one-out analysis for 9 GM taxa on IVDD.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Geng, Z., Wang, J., Chen, G. et al. Gut microbiota and intervertebral disc degeneration: a bidirectional two-sample Mendelian randomization study. J Orthop Surg Res 18, 601 (2023). https://doi.org/10.1186/s13018-023-04081-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-023-04081-0