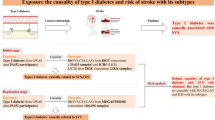
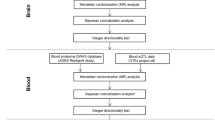
Abstract
Background
Stroke is a common neurological disorder that disproportionately affects middle-aged and elderly individuals, leading to significant disability and mortality. Recently, human blood metabolites have been discovered to be useful in unraveling the underlying biological mechanisms of neurological disorders. Therefore, we aimed to evaluate the causal relationship between human blood metabolites and susceptibility to stroke.
Methods
Summary data from genome-wide association studies (GWASs) of serum metabolites and stroke and its subtypes were obtained separately. A total of 486 serum metabolites were used as the exposure. Simultaneously, 11 different stroke phenotypes were set as the outcomes, including any stroke (AS), any ischemic stroke (AIS), large artery stroke (LAS), cardioembolic stroke (CES), small vessel stroke (SVS), lacunar stroke (LS), white matter hyperintensities (WMH), intracerebral hemorrhage (ICH), subarachnoid hemorrhage (SAH), transient ischemic attack (TIA), and brain microbleeds (BMB). A two‐sample Mendelian randomization (MR) study was conducted to investigate the causal effects of serum metabolites on stroke and its subtypes. The inverse variance-weighted MR analyses were conducted as causal estimates, accompanied by a series of sensitivity analyses to evaluate the robustness of the results. Furthermore, a reverse MR analysis was conducted to assess the potential for reverse causation. Additionally, metabolic pathway analysis was performed using the web-based MetOrigin.
Results
After correcting for the false discovery rate (FDR), MR analysis results revealed remarkable causative associations with 25 metabolites. Further sensitivity analyses confirmed that only four causative associations involving three specific metabolites passed all sensitivity tests, namely ADpSGEGDFXAEGGGVR* for AS (OR: 1.599, 95% CI 1.283–1.993, p = 2.92 × 10−5) and AIS (OR: 1.776, 95% CI 1.380–2.285, p = 8.05 × 10−6), 1-linoleoylglycerophosph-oethanolamine* for LAS (OR: 0.198, 95% CI 0.091–0.428, p = 3.92 × 10−5), and gamma-glutamylmethionine* for SAH (OR: 3.251, 95% CI 1.876–5.635, p = 2.66 × 10−5), thereby demonstrating a high degree of stability. Moreover, eight causative associations involving seven other metabolites passed both sensitivity tests and were considered robust. The association result of one metabolite (glutamate for LAS) was considered non-robust. As for the remaining metabolites, we speculate that they may potentially possess underlying causal relationships. Notably, no common metabolites emerged from the reverse MR analysis. Moreover, after FDR correction, metabolic pathway analysis identified 40 significant pathways across 11 stroke phenotypes.
Conclusions
The identified metabolites and their associated metabolic pathways are promising circulating metabolic biomarkers, holding potential for their application in stroke screening and preventive strategies within clinical settings.
Similar content being viewed by others
Introduction
Stroke is one of the most prevalent neurological disorders and is a major cause of disability and death among middle-aged and elderly individuals, posing a significant public health concern on a global scale [1]. According to the Global Burden of Disease estimation in 2019, stroke incidence was 12.2 million cases, the prevalent cases of stroke were 101 million, the number of disability-adjusted life-years was 143 million, and the number of deaths caused by stroke was 6.55 million[2]. Stroke has various subtypes, with ischemic stroke most commonly involved. Ischemic stroke can be further divided into three subtypes: large artery stroke (LAS), cardioembolic stroke (CES), and small vessel stroke (SVS) [3]. Furthermore, stroke includes intracerebral hemorrhage (ICH) and subarachnoid hemorrhage (SAH) [4]. Transient ischemic attack (TIA) is a robust predictor of stroke and is considered a minor stroke [5]. White matter hyperintensities (WMH) and brain microbleeds (BMB) are important risk factors for ischemic stroke [6] and ICH [7]. While the pathological processes vary among different stroke subtypes, they all involve the death of nerve cells [8]. Despite several studies on the nature of stroke, the biological mechanisms and risk factors underlying its occurrence remain unclear. Identifying modifiable risk factors for stroke is crucial for develo** preventative interventions.
Recently, the connection between metabolomics and stroke has gained attention. Metabolomics is used for biomarker discovery, providing insights into the processes of disease occurrence and progression by uncovering altered metabolic pathways and intermediate metabolites [9]. Metabolites are the end products or intermediate compounds in metabolism that provide essential functions in the human body. Multiple studies have demonstrated that metabolites are functional intermediates that can elucidate the potential biological mechanisms underlying disease genetics [26]. Among the 486 metabolites, 309 are known, while 177 are unknown. According to the Kyoto encyclopedia of genes and genomes (KEGG) database, the known metabolites can be assigned to eight broad metabolic categories: cofactors and vitamins, energy, lipids, nucleotides, peptides, amino acids, carbohydrates, and xenobiotics. Herein, we excluded 34 metabolite traits that could not be assigned IVs, leaving us with a subset of 452 serum metabolites for further analysis.
Instrumental variables selection
Herein, we selected SNPs with p-values below the locus-wide significance level (1 × 10–5) in the initial analysis as IVs to obtain comprehensive results and enhance sensitivity to IVs. Subsequently, all IVs underwent linkage disequilibrium (LD) clum** (r2 = 0.01; distance = 5000 kb) to mitigate the influence of correlated SNPs. Furthermore, Phenoscanner (http://www.phenoscanner.medschl.cam.ac.uk/) was screened to identify the potential pleiotropic effects. Additionally, we calculated the F-statistic [R2 (N–2)/(1–R2)], which assesses the strength of each instrument, where R2 represents the proportion of variance explained by the genetic instrument, and N is the effective sample size of GWAS [27]. The SNPs with an F-statistic threshold greater than ten were chosen for the subsequent MR analysis as they provided a reliable estimate of genetic variation [28]. Finally, we excluded palindromic SNPs [29] (where the effective allele is unclear) from our study.
Data sources on the stroke and its subtypes
Stroke is classified based on the clinical criteria defined by the World Health Organization (WHO) and the tenth edition of the International Classification of Diseases (ICD-10) [30]. Data for certain stroke subtypes were sourced from publicly available summary data provided by the MEGASTROKE consortium [14]. The MEGASTROKE consortium encompassed 446,696 individuals of European ancestry (40,585 any stroke (AS) cases and 406,111 controls). Within any ischemic stroke (AIS) category, there were 34,217 cases of overall AIS, 4,373 cases of LAS, 7,193 cases of CES, and 5,386 cases of SVS. Although the MEGASTROKE study included results for SVS, the cases were defined based on Trial of Org 10172 in Acute Stroke Treatment criteria [3] and did not specifically focus on MRI findings. Therefore, we conducted a study focusing on small vessel infarction using a sample of recent lacunar stroke (LS) cases, comprising 6,030 cases and 248,929 controls [31]. The WMH and BMB were imaging markers of cerebral microstructural damage [32]. The WMH is an increased brightness on T2-weighted brain images [33]. The BMBs are small, low-signal lesions identified on magnetic susceptibility-weighted imaging sequences or T2-weighted gradient-recalled echo sequences [34]. The summary data for WMH (N = 32,114) were derived from an expanded set of a recent GWAS study of brain imaging phenotypes conducted by the UK Biobank [35]. For BMBs, we could not find a GWAS specifically focused on individuals of European ancestry. However, in a recent multi-ethnic GWAS study on BMBs [36], we identified 2889 cases of microbleeds among the remaining 23,032 individuals after excluding patients with dementia and stroke. Summary-level data for the remaining stroke subtypes were generated from the latest FinnGen R9 Biobank [37], which included 3,749 cases of ICH, 3289 cases of SAH, and 18,398 cases of TIA. Further information on the GWAS can accessed at (https://www.finngen.fi/en). The studies in these consortia obtained approval from local research ethics committees and institutional review boards, and all participants provided written informed consent. Table 1 shows the characteristics of summarized datasets for the stroke subtypes.
MR analysis
Herein, a two-sample MR analysis was utilized to evaluate the causal correlation between serum metabolites and stroke and its subtypes. Subsequently, the fixed-effects or random-effects IVW method was employed as the primary MR analysis. The choice between the fixed-effects and random-effects IVW methods depends on heterogeneity and pleiotropy. The fixed-effects IVW model estimates are given higher significance when neither heterogeneity nor pleiotropy exists. In cases of heterogeneity without pleiotropy, we favor the random-effects IVW model. The random-effects IVW method is chosen for its ability to provide unbiased estimates by accounting for potential horizontal pleiotropy and striving to achieve balance in this context [38]. To enhance the robustness of results, we employed the MR-Egger method, weighted median analysis, and MR pleiotropy residual sum and outlier (MR-PRESSO) test as sensitivity analysis methods. The MR-Egger method considers directional horizontal pleiotropic effects. Whenever the intercept term significantly deviates from zero, it indicates the presence of invalid instruments and suggests potential bias in the IVW method [39]. The I2 value and Q-test assessed the potential heterogeneity and identified outliers in the IVW and MR-Egger analyses. The weighted median analysis requires at least half of the instruments to be valid, and the final overall MR estimate is determined by taking the median of causal estimates from each SNP [40]. The MR-PRESSO test was also conducted to identify potential horizontal pleiotropy and correct for its impact by removing outliers [41]. Additionally, we performed leave-one-out analyses to further evaluate the robustness of associations observed by individual SNP drivers.
Herein, a p-value less than 0.05 was considered a nominal association. False Discovery Rate (FDR) correction was employed to control for false positives in multiple tests [42]. Associations were considered statistically significant if the estimated causal effect of a given metabolite had an FDR value of < 0.05. The statistical power (> 80%) was estimated using the mRnd power calculator (http://cnsgenomics.com/shiny/mRnd/) [11, 46]. Blood is the most used sample source for metabolomics identification because it contains numerous detectable metabolites and can be easily obtained in large sample sizes, facilitating the screening of circulating biomarkers for stroke risk [81]. Considering cholesterol is also present in various stroke outcomes, we can infer that cholesterol may influence the occurrence and progression of stroke by affecting bile acid metabolism. Additionally, the metabolites involved in steroid degradation include cholesterol, and this pathway is associated with multiple stroke outcomes. Therefore, we hypothesize that cholesterol may affect steroid degradation, thereby influencing stroke occurrence. The pathways of glutathione metabolism, pantothenate, and CoA biosynthesis, arginine biosynthesis, aminoacyl-tRNA biosynthesis, and alanine, aspartate, and glutamate metabolism are also present in multiple stroke outcomes involving the metabolites glutamate and aspartate. Aspartate remains stable in our results, allowing us to infer its influence on the mentioned pathways and its impact on stroke progression. However, glutamate is non-robust in our results, indicating the need for further research to validate it.
Additionally, there are differences between the metabolites primarily involved in pathway analysis and those in our significant results. Therefore, there may be other unexplored metabolic pathways. Further research is required to better understand the relationships between metabolites, metabolic pathways, and stroke outcomes. Moreover, we have also discovered the involvement of certain novel pathways in stroke pathogenesis, including “styrene degradation” playing a significant role in ICH and “clavulanic acid biosynthesis” in SAH. The specific mechanisms behind these findings also require further investigation.
The use of drugs can influence changes in the metabolite profile. For example, statin medications lead to extensive lipid alterations and effectively reduce cholesterol levels [82]. Recently, develo** neuroprotective peptide drugs has influenced the occurrence of stroke by affecting the metabolism of amino acids in the human body, including glutamate and aspartate [83]. Additionally, some cardiovascular drugs can influence P-glycoprotein, which regulates the absorption and excretion of xenobiotics. The P-glycoprotein is associated with ischemic stroke in mouse models [84]. Therefore, the use of drugs in stroke patients may interfere with measuring metabolites, emphasizing a challenge for specific research on the impact of a particular metabolite on stroke in the future.
Based on our findings and cross-validation with RCT trial results, this provides early predictive factors for future research on the utility of these biomarkers in blood tests for stroke prevention. This study suggests that targeting certain metabolites may be a promising area for future medication development in treating stroke.
Our study has several strengths. First, a major strength of this study lies in its extensive coverage of genetic variables to comprehensively analyze the genetically determined relation between blood metabolites and the eleven stroke phenotypes. Meanwhile, the genome-wide dataset for stroke subtypes genetic variables primarily utilized populations of European ancestry to mitigate potential biases arising from population differences. Second, using bidirectional MR designs largely avoided reverse causation and residual confounding. Third, applying the largest available dataset on various stroke subtypes in the field, along with extensive sensitivity analyses, ensured the robustness of our findings.
Nevertheless, this study has certain limitations that should be acknowledged. First, we leveraged exposure-specific GWAS data and outcomes from publicly available summary data, with potential sample overlaps that might introduce confounding biases. Additionally, the distinct data sources in this study may correspond to different population groups. These samples could exhibit substantial variations in population characteristics, including age, gender, and socioeconomic background. Such distinctions can potentially influence the interpretation of causal estimates and the validity of causal inferences. Second, owing to the relatively limited number of participants in the exposure dataset and the restricted range of metabolite types, some associations between different metabolites and stroke might be missing. Third, the study participants primarily consisted of individuals of European descent, necessitating an assessment of the generalizability of our findings to other populations. Fourth, to enhance the reliability of our findings, we employed multiple correction analyses. However, this approach might overlook potential metabolites causally related to stroke. Fifth, some metabolites and metabolic pathways covered in this study have not been fully elucidated regarding their functions and mechanisms in diseases, which limits our interpretation of the MR analysis results. Lastly, due to the limited variance explained by SNPs or sample size constraints in GWAS results, some of our MR analyses might lack sufficient power to detect small effects. Future investigations utilizing larger GWAS datasets promise to provide enhanced statistical power and more precise assessments of the genetic influences on metabolites. Although MR has assisted in identifying blood metabolites associated with stroke, there remains a need for prospective studies to delve into their potential mechanisms.
Conclusion
This two-sample MR study revealed the significant role of serum metabolites in the risk of 11 stroke subtypes. Identifying 28 remarkable causal associations between 25 metabolites and 9 stroke phenotypes, 40 significant metabolic pathways in 11 stroke phenotypes, and nominal causal associations of other metabolites contribute to our understanding of the intricate interplay between metabolites and the brain in the development of stroke. Moreover, they offer valuable potential as circulating metabolic biomarkers, holding promise for their application in stroke screening and preventive strategies within clinical settings. These findings contribute to the understanding of biological mechanisms underlying stroke and pave the way for future exploration of targeted therapeutic interventions.
Availability of data and materials
As the current study used published GWAS summary data, so, the relevant data is publically available. Further inquiries can be directed to the corresponding authors.
References
O’Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016;388(10046):761–75.
GBD 2019 Stroke Collaborators. Global, regional, and national burden of stroke and its risk factors, 1990–2019: a systematic analysis for the Global burden of disease study 2019. Lancet Neurol. 2021;20(10):795–820.
Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute stroke treatment. Stroke. 1993;24(1):35–41.
Ohashi SN, DeLong JH, Kozberg MG, Mazur-Hart DJ, van Veluw SJ, Alkayed NJ, et al. Role of inflammatory processes in hemorrhagic stroke. Stroke. 2023;54(2):605–19.
Amin HP, Madsen TE, Bravata DM, Wira CR, Johnston SC, Ashcraft S, et al. Diagnosis, workup, risk reduction of transient ischemic attack in the emergency department setting: a scientific statement from the American heart association. Stroke. 2023;54(3):e109–21.
Yang Y, Knol MJ, Wang R, Mishra A, Liu D, Luciano M, et al. Epigenetic and integrative cross-omics analyses of cerebral white matter hyperintensities on MRI. Brain J Neurol. 2023;146(2):492–506.
Lovelock CE, Cordonnier C, Naka H, Al-Shahi Salman R, Sudlow CL, Sorimachi T, et al. Antithrombotic drug use, cerebral microbleeds, and intracerebral hemorrhage: a systematic review of published and unpublished studies. Stroke. 2010;41(6):1222–8.
Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A, et al. An updated definition of stroke for the 21st century: a statement for healthcare professionals from the American heart association/American stroke association. Stroke. 2013;44(7):2064–89.
Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–9.
**ao G, He Q, Liu L, Zhang T, Zhou M, Li X, et al. Causality of genetically determined metabolites on anxiety disorders: a two-sample Mendelian randomization study. J Transl Med. 2022;20(1):475.
Tiedt S, Brandmaier S, Kollmeier H, Duering M, Artati A, Adamski J, et al. Circulating metabolites differentiate acute ischemic stroke from stroke mimics. Ann Neurol. 2020;88(4):736–46.
Zoghi S, Abbasi A, Heravi FS, Somi MH, Nikniaz Z, Moaddab SY, et al. The gut microbiota and celiac disease: Pathophysiology, current perspective and new therapeutic approaches. Crit Rev Food Sci Nutr. 2022. https://doi.org/10.1080/10408398.2022.2121262.
Mishra A, Malik R, Hachiya T, Jürgenson T, Namba S, Posner DC, et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature. 2022;611(7934):115–23.
Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524–37.
Schiano C, Benincasa G, Franzese M, Della Mura N, Pane K, Salvatore M, et al. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol Ther. 2020;210: 107514.
Kettunen J, Demirkan A, Würtz P, Draisma HH, Haller T, Rawal R, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122.
Lotta LA, Pietzner M, Stewart ID, Wittemans LBL, Li C, Bonelli R, et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet. 2021;53(1):54–64.
Smith CJ, Sinnott-Armstrong N, Cichońska A, Julkunen H, Fauman EB, Würtz P, et al. Integrative analysis of metabolite GWAS illuminates the molecular basis of pleiotropy and genetic correlation. Elife. 2022;11:e79348.
Reith C, Landray M, Devereaux PJ, Bosch J, Granger CB, Baigent C, et al. Randomized clinical trials–removing unnecessary obstacles. N Engl J Med. 2013;369(11):1061–5.
Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review. Res Synth Methods. 2019;10(4):486–96.
Burgess S, Timpson NJ, Ebrahim S, Davey SG. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. 2015;44(2):379–88.
Wang RZ, Huang SY, Li HQ, Yang YX, Chen SD, Yu JT. Genetic determinants of circulating metabolites and the risk of stroke and its subtypes. Eur J Neurol. 2022;29(12):3711–9.
Guo MN, Hao XY, Tian J, Wang YC, Li JD, Fan Y, et al. Human blood metabolites and lacunar stroke: A Mendelian randomization study. Int J Stroke. 2023;18(1):109–16.
Harshfield EL, Markus HS. Association of baseline metabolomic profiles with incident stroke and dementia and with imaging markers of cerebral small vessel disease. Neurology. 2023;101(5):e489–501.
Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ Clinresearch ed). 2021;375: n2233.
Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–50.
Kwok MK, Kawachi I, Rehkopf D, Schooling CM. The role of cortisol in ischemic heart disease, ischemic stroke, type 2 diabetes, and cardiovascular disease risk factors: a bi-directional Mendelian randomization study. BMC Med. 2020;18(1):363.
Bottigliengo D, Foco L, Seibler P, Klein C, König IR, Del Greco MF. A Mendelian randomization study investigating the causal role of inflammation on Parkinson’s disease. Brain J Neurol. 2022;145(10):3444–53.
Flatby HM, Ravi A, Damås JK, Solligård E, Rogne T. Circulating levels of micronutrients and risk of infections: a Mendelian randomization study. BMC Med. 2023;21(1):84.
Thayabaranathan T, Kim J, Cadilhac DA, Thrift AG, Donnan GA, Howard G, et al. Global stroke statistics 2022. Int J Stroke. 2022;17(9):946–56.
Traylor M, Persyn E, Tomppo L, Klasson S, Abedi V, Bakker MK, et al. Genetic basis of lacunar stroke: a pooled analysis of individual patient data and genome-wide association studies. The Lancet Neurology. 2021;20(5):351–61.
Debette S, Schilling S, Duperron MG, Larsson SC, Markus HS. Clinical significance of magnetic resonance imaging markers of vascular brain injury: a systematic review and meta-analysis. JAMA Neurol. 2019;76(1):81–94.
Jiang L, Cai X, Yao D, **g J, Mei L, Yang Y, et al. Association of inflammatory markers with cerebral small vessel disease in community-based population. J Neuroinflammation. 2022;19(1):106.
Greenberg SM, Vernooij MW, Cordonnier C, Viswanathan A, Al-Shahi Salman R, Warach S, et al. Cerebral microbleeds: a guide to detection and interpretation. The Lancet Neurology. 2009;8(2):165–74.
Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24(5):737–45.
Knol MJ, Lu D, Traylor M, Adams HHH, Romero JRJ, Smith AV, et al. Association of common genetic variants with brain microbleeds: a genome-wide association study. Neurology. 2020;95(24):e3331–43.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–18.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8.
Huang SY, Yang YX, Kuo K, Li HQ, Shen XN, Chen SD, et al. Herpesvirus infections and Alzheimer’s disease: a Mendelian randomization study. Alzheimer’s Res Ther. 2021;13(1):158.
Zhao J, Chen H, Zhuo C, **a S. Cannabis use and the risk of cardiovascular diseases: a mendelian randomization study. Front Cardiovasc Med. 2021;8: 676850.
Yu G, Xu C, Zhang D, Ju F, Ni Y. MetOrigin: Discriminating the origins of microbial metabolites for integrative analysis of the gut microbiome and metabolome. imeta. 2022;1(1):e10.
Gu Y, ** Q, Hu J, Wang X, Yu W, Wang Z, et al. Causality of genetically determined metabolites and metabolic pathways on osteoarthritis: a two-sample mendelian randomization study. J Transl Med. 2023;21(1):357.
Jiang Z, Sun J, Liang Q, Cai Y, Li S, Huang Y, et al. A metabonomic approach applied to predict patients with cerebral infarction. Talanta. 2011;84(2):298–304.
Guo W, Wang Y, Fan M, **e S, Zhao H, Wang J, et al. Integrating metabolomics and network pharmacology to explore the protective effect of gross saponins of Tribulus terrestris L. fruit against ischemic stroke in rat. J Ethnopharmacol. 2020;263:113202.
Zhang R, Meng J, Wang X, Pu L, Zhao T, Huang Y, et al. Metabolomics of ischemic stroke: insights into risk prediction and mechanisms. Metab Brain Dis. 2022;37(7):2163–80.
Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20(1):122–8.
Huang J, Zhao B, Weinstein SJ, Albanes D, Mondul AM. Metabolomic profile of prostate cancer-specific survival among 1812 Finnish men. BMC Med. 2022;20(1):362.
Yang W, Kim CK, Kim DY, Jeong HG, Lee SH. Gamma-glutamyl transferase predicts future stroke: a Korean nationwide study. Ann Neurol. 2018;83(2):375–86.
Bae ON, Serfozo K, Baek SH, Lee KY, Dorrance A, Rumbeiha W, et al. Safety and efficacy evaluation of carnosine, an endogenous neuroprotective agent for ischemic stroke. Stroke. 2013;44(1):205–12.
Au A. Metabolomics and lipidomics of ischemic stroke. Adv Clin Chem. 2018;85:31–69.
Wang H, Chen S, Han Z, Li T, Ma J, Chen X, et al. Screening of phospholipids in plasma of large-artery atherosclerotic and cardioembolic stroke patients with hydrophilic interaction chromatography-mass spectrometry. Front Mol Biosci. 2022;9: 794057.
Li T, Peng R, Wang F, Hua L, Liu S, Han Z, et al. Lysophosphatidic acid promotes thrombus stability by inducing rapid formation of neutrophil extracellular traps: a new mechanism of thrombosis. J Thromb Haemost. 2020;18(8):1952–64.
Ke C, Pan CW, Zhang Y, Zhu X, Zhang Y. Metabolomics facilitates the discovery of metabolic biomarkers and pathways for ischemic stroke: a systematic review. Metabol Off J Metabol Soc. 2019;15(12):152.
Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, Ke LY. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci. 2019;20(5):1149.
Pérez-Matos MC, Morales-Álvarez MC, Toloza FJK, Ricardo-Silgado ML, Mantilla-Rivas JO, Pinzón-Cortes JA, et al. The phospholipid linoleoylglycerophosphocholine as a biomarker of directly measured insulin resistance. Diabetes Metab J. 2017;41(6):466–73.
Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care. 2020;43(6):1319–25.
Georgakis MK, Harshfield EL, Malik R, Franceschini N, Langenberg C, Wareham NJ, et al. Diabetes mellitus, glycemic traits, and cerebrovascular disease: a mendelian randomization study. Neurology. 2021;96(13):e1732–42.
Zierer J, Kastenmüller G, Suhre K, Gieger C, Codd V, Tsai PC, et al. Metabolomics profiling reveals novel markers for leukocyte telomere length. Aging. 2016;8(1):77–94.
Collino S, Montoliu I, Martin FP, Scherer M, Mari D, Salvioli S, et al. Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS ONE. 2013;8(3): e56564.
Chou PS, Yang IH, Kuo CM, Wu MN, Lin TC, Fong YO, et al. The prognostic biomarkers of plasma trimethylamine N-oxide and short-chain fatty acids for recanalization therapy in acute ischemic stroke. Int J Mol Sci. 2023;24(13):10796.
Park Y, Park S, Yi H, Kim HY, Kang SJ, Kim J, et al. Low level of n-3 polyunsaturated fatty acids in erythrocytes is a risk factor for both acute ischemic and hemorrhagic stroke in Koreans. Nutr Res. 2009;29(12):825–30.
Abdelhamid AS, Brown TJ, Brainard JS, Biswas P, Thorpe GC, Moore HJ, et al. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev. 2018;7(7):Cd003177.
Qian S, You S, Sun Y, Wu Q, Wang X, Tang W, et al. Remnant cholesterol and common carotid artery intima-media thickness in patients with ischemic stroke. Circ Cardiovasc Imaging. 2021;14(4): e010953.
Kimberly WT, Wang Y, Pham L, Furie KL, Gerszten RE. Metabolite profiling identifies a branched chain amino acid signature in acute cardioembolic stroke. Stroke. 2013;44(5):1389–95.
Shen Z, **ang M, Chen C, Ding F, Wang Y, Shang C, et al. Glutamate excitotoxicity: Potential therapeutic target for ischemic stroke. Biomed Pharmacother Biomed Pharmacother. 2022;151:113125.
Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015;14:6.
Ishida T, Inoue T, Niizuma K, Konno N, Suzuki C, Inoue T, et al. Prediction of functional outcome in patients with acute stroke by measuring tRNA derivatives. Cerebrovasc Dis. 2020;49(6):639–46.
Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic modifications modulate normal and pathological functions in CNS. Transl Stroke Res. 2022;13(1):1–11.
Servillo L, Giovane A, Casale R, Cautela D, D’Onofrio N, Balestrieri ML, et al. Homostachydrine (pipecolic acid betaine) as authentication marker of roasted blends of Coffea arabica and Coffea canephora (Robusta) beans. Food Chem. 2016;205:52–7.
Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25(1):27–42.
Cao Y, Su N, Zhang D, Zhou L, Yao M, Zhang S, et al. Correlation between total homocysteine and cerebral small vessel disease: a mendelian randomization study. Eur J Neurol. 2021;28(6):1931–8.
Zhou P, Zhou L, Shi Y, Li Z, Liu L, Zuo L, et al. Neuroprotective effects of danshen chuanxiongqin injection against ischemic stroke: metabolomic insights by UHPLC-Q-orbitrap hrms analysis. Front Mol Biosci. 2021;8: 630291.
Jiang W, Gong L, Liu F, Ren Y, Mu J. Alteration of Gut Microbiome and Correlated Lipid Metabolism in Post-Stroke Depression. Front Cell Infect Microbiol. 2021;11: 663967.
Ren JX, Li C, Yan XL, Qu Y, Yang Y, Guo ZN. Crosstalk between oxidative stress and ferroptosis/oxytosis in ischemic stroke: possible targets and molecular mechanisms. Oxid Med Cell Longev. 2021;2021:6643382.
Wang X, Zhang L, Sun W, Pei LL, Tian M, Liang J, et al. Changes of metabolites in acute ischemic stroke and its subtypes. Front Neurosci. 2020;14: 580929.
Jia J, Zhang H, Liang X, Dai Y, Liu L, Tan K, et al. Application of metabolomics to the discovery of biomarkers for ischemic stroke in the murine model: a comparison with the clinical results. Mol Neurobiol. 2021;58(12):6415–26.
Jiang W, Chen J, Gong L, Liu F, Zhao H, Mu J. Alteration of glycerophospholipid metabolism in hippocampus of post-stroke depression rats. Neurochem Res. 2022;47(7):2052–63.
Charach G, Karniel E, Novikov I, Galin L, Vons S, Grosskopf I, et al. Reduced bile acid excretion is an independent risk factor for stroke and mortality: a prospective follow-up study. Atherosclerosis. 2020;293:79–85.
Würtz P, Wang Q, Soininen P, Kangas AJ, Fatemifar G, Tynkkynen T, et al. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol. 2016;67(10):1200–10.
Dergunova LV, Filippenkov IB, Limborska SA, Myasoedov NF. Neuroprotective peptides and new strategies for ischemic stroke drug discoveries. Genes. 2023;14(5):953.
DeMars KM, Yang C, Hawkins KE, McCrea AO, Siwarski DM, Candelario-Jalil E. Spatiotemporal changes in P-glycoprotein levels in brain and peripheral tissues following ischemic stroke in rats. J Exp Neurosci. 2017;11:1179069517701741.
Acknowledgements
The authors offer their thanks and appreciation to the MEGASTROKE Consortium, the International Stroke Genetics consortia, the UK Biobank, FinnGen, Twins UK and KORA Study. We thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
Funding
This study was not supported by fund.
Author information
Authors and Affiliations
Contributions
TZ and GL designed the study, contributed to the data analysis, and wrote the manuscript. JZ and YC contributed to the data collection and data analysis. YC, JY and GL contributed to manuscript writing and revision of the manuscript. YC and GL helped us with drawing. All authors read and approved the final draft of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Here, our study is based on the large-scale GWAS datasets, and not the individual-level data. The studies included in these consortia obtained approval from local research ethics committees and institutional review boards, and all participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
STROBE-MR checklist of recommended items to address in reports of Mendelian randomization studies.
Additional file 2:
The result for leave-one-out analysis.
Additional file 3: Table S1
. The Results of all stroke phenotypes.
Additional file 4: Table S2
. The Results of all stroke phenotypes (P < 0.05).
Additional file 5: Table S3
. The reverse results of all stroke phenotypes (P < 0.05).
Additional file 6: Table S4
. The results of significant metabolites, metabolic pathways, power, and metabolite-related features.
Additional file 7: Table S5
. The relevant SNP results for all stroke phenotypes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, T., Cao, Y., Zhao, J. et al. Assessing the causal effect of genetically predicted metabolites and metabolic pathways on stroke. J Transl Med 21, 822 (2023). https://doi.org/10.1186/s12967-023-04677-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-023-04677-4