Abstract
Gene expression and posttranscriptional regulation can be strongly influenced by epigenetic modifications. N6-methyladenosine, the most extensive RNA modification, has been revealed to participate in many human diseases. Recently, the role of RNA epigenetic modifications in the pathophysiological mechanism of female reproductive diseases has been intensively studied. RNA m6A modification is involved in oogenesis, embryonic growth, and foetal development, as well as preeclampsia, miscarriage, endometriosis and adenomyosis, polycystic ovary syndrome, premature ovarian failure, and common gynaecological tumours such as cervical cancer, endometrial cancer, and ovarian cancer. In this review, we provide a summary of the research results of m6A on the female reproductive biology and pathophysiology in recent years and aim to discuss future research directions and clinical applications of m6A-related targets. Hopefully, this review will add to our understanding of the cellular mechanisms, diagnostic biomarkers, and underlying therapeutic strategies of female reproductive system diseases.
Video Abstract
Similar content being viewed by others
Background
As a result of the growing interest in understanding how DNA and RNA modifications function, epigenomics has become a cutting-edge field [1]. Scientists have identified numerous layers of epigenetic modulation that are derived from the modification of DNA and proteins; however, RNA modification remains elusive [2]. Over 160 RNA chemical modifications have been discovered thus far [3]. The multitude of RNA modifications has added a new level of complexity to gene regulation. In eukaryotic mRNA, m6A RNA modification is the most abundant modification, which is defined as the methylation of adenosine 6 positions in mRNA and some noncoding RNA. N6-methyladenosine has been proven to be connected with various metabolic and physiologic processes, such as RNA transcript splicing, translation efficiency, nuclear export, stability, and decay [4]. m6A shares the same base pairing with unmodified adenosine, which prevents it from being detected by conventional sequencing or hybridization methods. A mystery surrounded the transcriptome distribution of m6A until a combination of m6A-specific methylated RNA immunoprecipitation and next-generation sequencing was devised. By employing these methods, scientists were able to determine that m6A residues were located in evolutionarily conserved regions within humans and mice. m6A residues were found to be contained primarily within internal mRNA sequences and appeared in the poly A tail, especially in the 3'UTR of mRNAs near the stop codon, and they always share a consensus motif of RRACH (R = A/G, H = A/C/U), and a terminal U is dominant part of the consensus [5,6,7,8].
The process of RNA methylation is controlled by methyltransferases, demethylases, and recognition factors. A methyltransferase and a demethylase catalyse the methylation of RNA dynamically, and the m6A function is determined by its recognition factor. Regulators may also be referred to as ''writers'', ''erasers'', and ''readers''. Although these regulators have been identified, the specific function of this process has not been well established. Cell types and environmental conditions may affect the expression level and biological function of m6A effectors. Furthermore, these factors can also be influenced by RNA species, abundances, secondary structures, intracellular location, translation state, and other heterogeneous factors. Taking these factors into account, it may be difficult to explain the exact function of m6A [9].
The normal physiological functions of the female reproductive system depend on the gonadal axis of the reproductive endocrine system, which regulates ovulation and the menstrual cycle. The maintenance of normal pregnancy depends on the establishment of endometrial receptivity, normal embryogenesis, and the formation of an immune microenvironment at the maternal-foetal interface. Several studies have shown that m6A modification is related to the regulation of physiological functions of the female reproductive system [10]. For example, by controlling the translation of Pgr mRNA through m6A modification, METTL3 is crucial for efficient P4 signaling during embryogenesis. m6A‑RNA immunoprecipitation‑qPCR showed that Pgr mRNA transcript was a target for METTL3-dependent m6A mRNA methylation [11]. In the process of ovarian aging, researchers found that the ovarian aging can be alleviated by up-regulating the FTO level in ovarian granulosa cells to reduce the m6A level of FOS-mRNA-3′UTR [12].However, the subtle mechanism of m6A modification involved in the physiological function of the female reproductive system remains to be elucidated.
Common disorders of the female reproductive system include benign diseases, such as inflammatory diseases of the reproductive organs, infertility, endometriosis, and polycystic ovarian syndrome; complications of pregnancy, such as miscarriage, preeclampsia, and gestational diabetes mellitus; and some malignant diseases, such as cervical cancer, endometrial cancer, and ovarian cancer. In recent years, it has been found that epigenetic abnormalities may be associated with the development of several female reproductive disorders. For example, epigenetic modifications such as DNA methylation, histone methylation and acetylation and long noncoding RNAs are involved in regulating follicular granulosa cell proliferation, endometrial receptivity and decidualization in early pregnancy, which affect ovulation, embryo implantation, and labour initiation [13]. Abnormalities in these epigenetic modifications may be associated with polycystic ovary syndrome, infertility due to repeated embryo implantation failure, and pregnancy complications. Some patients with endometriosis may be born with congenital epigenetic molecular abnormalities, and the recurrent bleeding and repair processes of endometriosis lesions may also trigger acquired epigenetic events [14]. In addition, some chemical toxicants, such as Bisphenol A (BPA), can affect ovarian function, embryonic development, and gamete quality during fertilization through epigenetic modifications such as CpG island methylation, histone modifications, and noncoding RNA production [15].
The function of epigenetic modification in the female reproductive system has received increasing attention over the last few years. Despite this, research on m6A modification and the female reproductive system is still in its infancy. A comprehensive review of recent research advances in m6A modification and its roles in pathogenesis, diagnosis, and molecular targeted therapies for gynaecological reproductive disease is presented in this paper. We discuss the potential directions of m6A modification for future research in obstetrical and gynaecological disorders, aiming to clarify the role played by RNA methylation in female reproductive system diseases and malignant tumours. In doing so, we can gain a novel perspective on these diseases.
The mechanism and regulation of m6A RNA methylation
m6A writers
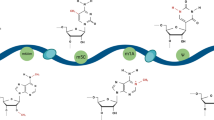
An N6-adenosine methyltransferase complex (MTC) is required for the installation of this modification (Fig. 1). The MTC comprises the METTL3 catalytic subunit, as well as the METTL14, WTAP, VIRMA, RBM15, and ZC3H13 accessory subunits. A 70 kDa protein named METTL3 was the first known m6A writer [3, 16]. The METTL3 subunit is a key component of the catalytic process, facilitating the reception of methyl groups from the SAM moiety by RNA adenine. METTL14, a molecular homologue of METTL3, was discovered by researchers as a new methyltransferase [17]. When METTL14 and METTL3 combine, a stable heterodimer is formed. It is believed that this complex mediates m6A deposition on nuclear RNA in mammals. A structural requirement for the activation of METTL3 is METTL14. METTL14 acts as an essential component to facilitate RNA recognition and binding. Although METTL14 is not directly involved in catalysis, it stabilizes the conformation of METTL3 to enhance catalytic activity [18, 19]. By decreasing METTL14 expression, methylated RNA binding and the splicing-related protein DGCR8, which plays an important role in maturing RNA, were suppressed [20].
Detailed mechanism of m6A readers, writers, and erasers. N6-methyladenosine is methylated by the methyltransferase complex, which contains METTL3, METTL14, WTAP, KIAA1429, and RBM15/15B. The m6A erasers FTO and ALKBH5 can reversibly remove m6A. The reader is responsible for performing the biological functions of m6A, such as RNA translation, decay, splicing, and translation
RNA-binding domains in RBM15 and RBM15B enable WTAP–METTL3 to bind to specific mRNAs and determine the methylated m6A consensus sequences. As part of the m6A methylation complex, RBM15/15B binds to XIST, and X-chromosome genes can be silenced by this transcription factor [21]. Researchers have identified that WTAP contains no obvious catalytic domains, but it initiates the localization of METTL3-METTL14 in the nuclear speckle and facilitates catalysis. Both WTAP and METTL3 regulate transcription and RNA processing genes by regulating their expression and alternative splicing. Morpholino-mediated knockdown of WTAP or METTL3 in zebrafish embryos resulted in defects in tissue differentiation and an increase in apoptosis [23]. In HeLa cells, KIAA1429 mediates the preferred deposition of m6A in the 3'UTR near the stop codon and contributes to APA [24]. Furthermore, its homologue, Virilizer, is essential for the alternative splicing of the sex determination factor in Drosophila [25].
Due to METTL16’s role in regulating SAM homeostasis, it has been linked to several modifications in the epitranscriptome of m6A [26]. Scientists have investigated whether METTL16 methylates MAT2A (encoding SAM synthetase) and U6 snRNA [82]. Table 1 summarizes the physiological functions of m6A RNA methylation in the female reproductive system.
m6A RNA methylation in pregnancy and female reproductive disorder
Preeclampsia and gestational diabetes mellitus
Research has found that placental trophoblasts of preeclampsia patients had significantly higher levels of m6A, METTL3, and HNRNPC1/C2 [83]. A recent study demonstrated that METTL3 promotes m6A RNA methylation in the placenta of preeclamptic patients, resulting in a change from pre-miRNA 497-5p/145-5p to mature miRNA 497-5p/145-5p [84]. In preeclampsia samples, the levels of several heat shock proteins were elevated, and the peak of m6A was primarily located in the CDS region near the 3'UTR. Researchers have discovered that the m6A readers IGF2BPs can enhance gene expression by stabilizing mRNA when bound to mRNA [140,141,142]. Although research on these chemical modifications is not as intensive as that on m6A, the role of these chemical modifications in female reproductive disorders deserves attention and has great therapeutic potential.
m5C is a conserved and common RNA modification. It is primarily found in eukaryotic tRNAs and rRNAs [143]. In an animal experiment, deletion of the m5C methyltransferase Nsun5 resulted in suppression of ovarian function and embryonic development arrest in mice [144]. In several bioinformatics studies on ovarian and endometrial cancers, m5C-related genes were closely associated with tumour chemotherapy sensitivity and overall survival [145,146,147]. In addition, abnormal m5C modification may be involved in TRDMT1-mediated granulosa cell death, which in turn causes premature ovarian failure [148]. Another study in Drosophila confirmed that YPS promotes the proliferation and differentiation of germinal stem cells in the Drosophila ovary by binding RNA containing m5C modifications [149]. However, the role of m5C in human ovarian function remains to be further investigated.
m1A and m7G modifications are also present in eukaryotic mRNAs [150]. m1A modifications may produce their biological effects by enhancing RNA‒protein interactions or by altering RNA secondary structures [151]. m7G modification is usually catalysed by METTL1 and located at the 5’ caps and internal positions of eukaryotic mRNA [152]. Several studies on endometrial cancer have found that m1A- and m7G-related lncRNAs and miRNAs can be used to construct tumour-related prognostic models and are associated with different immune infiltration phenotypes and drug susceptibility in endometrial cancer [153,154,155,156]. Pseudouridine is the most abundant modified nucleotide in RNA [157]. In a study on ovarian cancer, a pseudouridine synthase, PUS7, was considered a potential diagnostic marker for ovarian cancer [158].
m6A RNA methylation sheds light on the diagnosis and treatment of several diseases
The TNM stage of gastric cancer is associated with METTL14 expression. According to an overall survival analysis, overall survival rates were higher for patients with higher levels of METTL14 expression. Based on multivariate analysis, METTL14 expression levels were associated with improved outcomes [159]. Non-small cell lung carcinoma cells undergo drug resistance and metastasis via diverse pathways when METTL3 increases m6A modification of both YAP and lncRNA MALAT1 [160]. FTO promotes cell proliferation of acute myeloid leukaemia in an m6A-dependent manner. Two inhibitors of FTO have been successfully developed by Huang and colleagues. These inhibitors, FB23 and FB23-2, bind directly to FTO and inhibit its m6A demethylase activity. The inhibitors display significantly improved inhibitory activity on FTO demethylation of m6A-RNA in vitro. Using these inhibitors, researchers inhibited the proliferation of a panel of AML cell lines and primary AML leukaemia stem cells in patient-derived xenotransplantation mice. Therefore, FTO and its analogues may be effective molecular targets for inhibiting leukaemogenesis in leukaemia stem cells [161,162,163]. Previous studies also indicated that YTHDF1 was strongly associated with a poor prognosis for ovarian cancer, breast cancer, and other female reproductive disorders [132, 164, 165]. The low expression of KIAA1429, an m6A methyltransferase, significantly increased dendritic cell infiltration. Researchers have shown that KIAA1429 expression affects immune checkpoint blockade therapeutic efficacy and boosts intratumoral antitumour immunity. Patients with low KIAA1429 expression experienced survival benefits from anti-PD-L1 immunotherapy [166,167,168].
It has been demonstrated that m6A regulators are associated with the prognosis of cervical cancer patients. m6A methylation regulators may be key regulators of PD-L1 expression and immune cell infiltration and may have an important impact on the TIME of cervical cancer [169]. In recent years, the role of m6A modification in PARP resistance in ovarian cancer has attracted the interest of several scientists. In BRCA-mutated ovarian cancer cells, m6A modification of FZD10 mRNA promoted PARP resistance through upregulation of the Wnt/β-catenin pathway [170]. Another study on olaparib resistance in ovarian cancer cells showed that m6A modifications of the olaparib pharmacogene were increased in resistant ovarian cancer cells [171]. These m6A modification sites could be used as potential pharmacoepitranscriptomics markers of drugs. Studies have confirmed that ALKBH5 expression is upregulated in cisplatin-resistant ovarian cancer and that overexpression of the ALKBH5-HOXA10 loop activates the JAK2/STAT3 signalling pathway, leading to chemoresistance in ovarian cancer [172]. In future studies, targeted blockade against the ALKBH5-HOXA10 loop may reverse chemoresistance in ovarian cancer cells. Based on the above studies, it is clear that m6A regulators are closely linked to clinical treatment and diagnosis.
Conclusion
The actions of m6A modification have gradually become apparent in recent years as high-throughput sequencing technologies and highly specific antibodies against m6A have been developed. In the field of m6A RNA methylation, great progress has been made in revealing potential mechanisms of the onset and progression of female reproductive system disorders. Differentially expressed m6A regulator genes are found in a large number of gynaecological cells compared to normal cells and serve as triggers in gynaecological disease progression. Additionally, imbalanced RNA m6A has also been identified in these diseases. To clarify the molecular mechanisms underlying certain refractory obstetrics and gynaecology diseases, such as endometriosis and preeclampsia, abnormal RNA m6A modification is an important research direction.
Various female reproductive cancers are known to be affected by m6A modification and its regulators. Crystal structure data, for example, have provided insight into cancer-associated mutations in METTL14. Mutations in METTL14 that cause an inefficient RNA-binding domain are found in endometrial cancers, and the mutated form of the enzyme shows partially reduced activity. It has also been found that m6A mRNA methylation modulates AKT activity, which is necessary for endometrial cancer proliferation and tumorigenicity [126]. By binding to a locus of the MYC gene, IGF2BP2 enhances the proliferation, metastasis, and aerobic glycolysis of cervical cancer cells [173]. These findings indicate that the role of m6A regulators in female reproductive organs is different based on cell type, therapeutic issues, and pathological conditions.
Clinical diagnostics and therapeutics can also target m6A regulators in disorders of the female reproductive system. The oncogenic mechanism of mRNA m6A modification in gynaecologic malignant tumorigenesis may become a direction for future research. m6A regulators and their upstream and downstream signalling pathways may provide certain methods to elucidate the pathophysiological mechanisms of gynaecologic malignant tumours and establish prognostic models. Modification of pharmacogene mRNAs may alter their pharmacokinetics and pharmacodynamics, thus affecting drug efficacy. In future studies, not only do the pathophysiological mechanisms of female reproductive system diseases in which m6A is involved deserve attention but also the m6A modifications occurring in the pharmacodynamic gene targets corresponding to drugs need to be considered.
For the successful design of tissue- or cell-specific RNA methylation agonists or inhibitors, more multicentre and large-scale studies are needed. Many essential issues must be addressed in a detailed manner. A very limited number of cell and animal models are used in all cited papers, and the results are hence subject to variation. First, although METTL4, FTO, and ALKBH5 are commonly known and well-studied m6A regulatory factors, the role of less studied m6A regulatory factors, such as RBM15/15B and ZC3H13, may only be the tip of the iceberg. Further research is needed on the interactions between RNA modification regulators and their downstream targets in the female reproductive system. Second, the reproductive-endocrine system plays a pivotal role in the progression of gynaecological diseases. Research currently focuses on the influence of m6A on cellular behaviour. The interplay between the hormonal milieu and endocrine system related to m6A modification remains a mystery and should be addressed in the future. Finally, the majority of published studies have concentrated only on the molecular mechanism of m6A modification. A greater focus should be placed on the diagnostic and therapeutic properties of m6A. According to a large body of evidence, m6A modifications are related to clinical phenotypes and prognoses. m6A is involved in the common pathological features shared by benign and malignant diseases of the female reproductive system, and different m6A phenotypes in these diseases may be involved in the transformation between benign and malignant diseases and between different pathological types in the same disease.
Compared with other reviews in this field, our article provides a more comprehensive overview of the relationship between m6A and the physiology and pathology of the female reproductive system. Whether it is germ cell genesis, embryonic development or benign and malignant diseases of the female reproductive system, we searched for appropriate literature to address them. In particular, we briefly outlined other RNA modifications. The connection between these RNA modifications and m6A modifications may become a new direction in future studies. In addition, we look forward to the involvement of m6A modifications in the diagnosis and treatment of female reproductive system diseases. m6A modifications deserve to be noticed for their clinical applications in female reproductive system diseases.
Undoubtedly, the pathogenesis of female reproductive system diseases is quite complex, and changes in the levels of m6A regulators dynamically regulate the level of m6A modifications in the female reproductive system, altering the abundance and function of related mRNAs and proteins and ultimately determining the onset of disease. However, the application of m6A regulators to clinical diagnosis and treatment requires improving the sensitivity and specificity of the relevant biomarkers. RNA m6A modification can likely be used to classify clinical phenotypes of several female reproductive disorders and to predict the mutual transformation of these disorders.
Availability of data and materials
Not applicable.
Abbreviations
- 3′ UTR:
-
3'-Untranslated region
- ALCAM:
-
Activated leukocyte cell adhesion molecule
- APA:
-
Alternative polyadenylation
- BAMBI:
-
BMP and activin membrane bound inhibitor
- BPDE:
-
Benzo(a)pyrene-trans-7,8-diol-9,10-epoxide
- CDS:
-
Sequence coding for aminoacids in protein
- CNOT1:
-
CCR4-NOT transcription complex subunit 1
- CRD:
-
Coding region instability determinant
- CREBBP:
-
CREB binding protein
- CTX:
-
Cytoxan
- CYR61:
-
Cysteine-rich 61
- DDT:
-
D-dopachrome tautomerase
- DGCR8:
-
DiGeorge syndrome critical region gene 8
- FBXW7:
-
F-box and WD repeat-containing 7
- FOXO3:
-
Forkhead box O3;
- GV:
-
Germinal vesicle
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein
- HOXB13:
-
Homeobox B13
- HSPA1:
-
Heat Shock Protein 70
- IGF1:
-
Insulin-like growth factor-1
- IGF2BP:
-
Insulin-like growth factor 2 mRNA-binding protein
- ITSN2:
-
Intersectin 2
- KDM3B:
-
Lysine demethylase 3B
- VIRMA:
-
Vir like m6A methyltransferase associated
- lncRNA:
-
Long non-coding RNA
- LRR:
-
Leucine-rich repeat
- m6A:
-
N6-methyladenosine
- m6Am:
-
N6,2′-O-dimethyladenosine
- MAT2A:
-
Methionine adenosyltransferase 2A
- METTL14:
-
Methyltransferase-like Protein 14
- MII:
-
Metaphase II
- MTC:
-
N6-adenosine methyltransferase complex
- MXD1:
-
MAX dimerization protein 1
- MYB:
-
MYB proto-oncogene, transcription factor
- MYC:
-
MYC proto-oncogene
- NRL:
-
Nucleosomal repeat length
- NXF1:
-
Nuclear RNA export factor 1
- OC:
-
Ovarian cancer
- OGN:
-
Osteoglycin
- PAPPA2:
-
Pappalysin 2
- PD-L1:
-
Programmed cell death-ligand 1
- PPARG:
-
Peroxisome proliferator activated receptor gamma
- pre-mRNA:
-
Pre-messenger RNA
- RBM15/15B:
-
RNA binding motif protein 15/15B
- SAM:
-
S-adenosylmethionine
- SMPD1:
-
Sphingomyelin phosphodiesterase 1
- SRSF3:
-
Serine and arginine rich splicing factor 3
- TNM:
-
Tumor node metastasis
- U6 snRNA:
-
U6 spliceosomal small nuclear RNA
- VCR:
-
Vertebrate conserved region
- WTAP:
-
Wilms tumor 1-associating protein
- XIST:
-
X-inactive specific transcript
- YTH family:
-
YT521-B homology domain family
- ZC3H13:
-
Zinc finger CCCH-type containing 13
References
Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. https://doi.org/10.1038/nrg3724.
Zhao LY, Song J, Liu Y, Song CX, Yi C. Map** the epigenetic modifications of DNA and RNA. Protein Cell. 2020;11(11):792–808. https://doi.org/10.1007/s13238-020-00733-7.
Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34(Database issue):D145–9. https://doi.org/10.1093/nar/gkj084.
Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications@@: form, distribution, and function. Science. 2016;352(6292):1408–12. https://doi.org/10.1126/science.aad8711.
Viegas IJ, de Macedo JP, Serra L, De Niz M, Temporão A, Silva Pereira S, et al. N(6)-methyladenosine in poly(A) tails stabilize VSG transcripts. Nature. 2022;604(7905):362–70. https://doi.org/10.1038/s41586-022-04544-0.
**ang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543(7646):573–6. https://doi.org/10.1038/nature21671.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. https://doi.org/10.1038/nature11112.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149(7):1635–46. https://doi.org/10.1016/j.cell.2012.05.003.
Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–50. https://doi.org/10.1016/j.molcel.2019.04.025.
Sun X, Lu J, Li H, Huang B. The role of m(6)A on female reproduction and fertility: from gonad development to ovarian aging. Front Cell Dev Biol. 2022;10:884295. https://doi.org/10.3389/fcell.2022.884295.
Zheng ZH, Zhang GL, Jiang RF, Hong YQ, Zhang QY, He JP, et al. METTL3 is essential for normal progesterone signaling during embryo implantation via m(6)A-mediated translation control of progesterone receptor. Proc Natl Acad Sci U S A. 2023;120(5):e2214684120. http://doi.org/https://doi.org/10.1073/pnas.2214684120
Jiang ZX, Wang YN, Li ZY, Dai ZH, He Y, Chu K, et al. The m6A mRNA demethylase FTO in granulosa cells retards FOS-dependent ovarian aging. Cell Death Dis. 2021;12(8):744. https://doi.org/10.1038/s41419-021-04016-9.
Liu H, Huang X, Mor G, Liao A. Epigenetic modifications working in the decidualization and endometrial receptivity. Cell Mol Life Sci. 2020;77(11):2091–101. https://doi.org/10.1007/s00018-019-03395-9.
Koninckx PR, Ussia A, Adamyan L, Wattiez A, Gomel V, Martin DC. Pathogenesis of endometriosis: the genetic/epigenetic theory. Fertil Steril. 2019;111(2):327–40. https://doi.org/10.1016/j.fertnstert.2018.10.013.
Chianese R, Troisi J, Richards S, Scafuro M, Fasano S, Guida M, et al. Bisphenol A in reproduction: epigenetic effects. Curr Med Chem. 2018;25(6):748–70. https://doi.org/10.2174/0929867324666171009121001.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187–200. https://doi.org/10.1016/j.cell.2017.05.045.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5. https://doi.org/10.1038/nchembio.1432.
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575–8. https://doi.org/10.1038/nature18298.
Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63(2):306–17. https://doi.org/10.1016/j.molcel.2016.05.041.
Zhang BY, Han L, Tang YF, Zhang GX, Fan XL, Zhang JJ, et al. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2020;24(12):7015–23. http://doi.org/https://doi.org/10.26355/eurrev_202006_21694
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–73. https://doi.org/10.1038/nature19342.
** X-L, Sun B-F, Wang L, **ao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–89. https://doi.org/10.1038/cr.2014.3.
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8(1):284–96. https://doi.org/10.1016/j.celrep.2014.05.048.
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. https://doi.org/10.1038/s41421-018-0019-0.
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–4. https://doi.org/10.1038/nature20577.
Ruszkowska A. METTL16, methyltransferase-like protein 16: current insights into structure and function. Int J Mol Sci. 2021;22(4):2176. https://doi.org/10.3390/ijms22042176.
Pendleton KE, Chen B, Liu K, Hunter OV, **e Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824-35.e14. https://doi.org/10.1016/j.cell.2017.05.003.
Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71(6):986-1000.e11. https://doi.org/10.1016/j.molcel.2018.08.004.
Chen PB, Shi GX, Liu T, Li B, Jiang SD, Zheng XF, et al. Oxidative stress aggravates apoptosis of nucleus pulposus cells through m(6)A modification of MAT2A Pre-mRNA by METTL16. Oxid Med Cell Longev. 2022;2022:4036274. https://doi.org/10.1155/2022/4036274.
Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol Cell. 2018;71(6):1001-11.e4. https://doi.org/10.1016/j.molcel.2018.07.025.
Zhao Y, Shi Y, Shen H, **e W. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13(1):35. https://doi.org/10.1186/s13045-020-00872-8.
Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285(19):14701–10. https://doi.org/10.1074/jbc.M110.104711.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. https://doi.org/10.1038/nature12730.
Du H, Zhao Y, He J, Zhang Y, ** H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. https://doi.org/10.1038/ncomms12626.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–28. https://doi.org/10.1038/cr.2017.15.
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017. https://doi.org/10.7554/eLife.31311.
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115–27. https://doi.org/10.1038/cr.2017.99.
Mao Y, Dong L, Liu X-M, Guo J, Ma H, Shen B, et al. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-13317-9.
Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19(2):1262–70. https://doi.org/10.1128/mcb.19.2.1262.
Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657–75. https://doi.org/10.1007/s00018-012-1186-z.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95. https://doi.org/10.1038/s41556-018-0045-z.
Doyle GAR, Leeds PF, Fleisig AJ, Ross J, Betz NA, Prokipcak RD. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 1998;26(22):5036–44. https://doi.org/10.1093/nar/26.22.5036.
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, et al. Regulation of co-transcriptional pre-mRNA splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell. 2019;76(1):70-81.e9. https://doi.org/10.1016/j.molcel.2019.07.005.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4. https://doi.org/10.1038/nature14234.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–308. https://doi.org/10.1016/j.cell.2015.08.011.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. https://doi.org/10.1126/science.1141634.
Zhao X, Yang Y, Sun BF, Shi Y, Yang X, **ao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–19. https://doi.org/10.1038/cr.2014.151.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7. https://doi.org/10.1038/nchembio.687.
Galganski L, Urbanek MO, Krzyzosiak WJ. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45(18):10350–68. https://doi.org/10.1093/nar/gkx759.
Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature. 2017;541(7637):371–5. https://doi.org/10.1038/nature21022.
Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 2017;45(19):11356–70. https://doi.org/10.1093/nar/gkx778.
Zou S, Toh JD, Wong KH, Gao YG, Hong W, Woon EC. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep. 2016;6:25677. https://doi.org/10.1038/srep25677.
Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, et al. Structure of human RNA N(6)-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42(7):4741–54. https://doi.org/10.1093/nar/gku085.
Heasman J. Patterning the early Xenopus embryo. Development. 2006;133(7):1205–17. https://doi.org/10.1242/dev.02304.
Qi ST, Ma JY, Wang ZB, Guo L, Hou Y, Sun QY. N6-methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J Biol Chem. 2016;291(44):23020–6. https://doi.org/10.1074/jbc.M116.748889.
Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137(6):859–70. https://doi.org/10.1242/dev.039487.
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14(5):e1007412. https://doi.org/10.1371/journal.pgen.1007412.
Qi Z, Liu Y, Yang H, Yang X, Wang H, Liu B, et al. Protective role of m(6)A binding protein YTHDC2 on CCNB2 in manganese-induced spermatogenesis dysfunction. Chem Biol Interact. 2022;351:109754. https://doi.org/10.1016/j.cbi.2021.109754.
Zhao X, Tian GG, Fang Q, Pei X, Wang Z, Wu J. Comparison of RNA m(6)A and DNA methylation profiles between mouse female germline stem cells and STO cells. Mol Ther Nucleic Acids. 2021;23:431–9. https://doi.org/10.1016/j.omtn.2020.11.020.
Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, et al. The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67(6):1059-67.e4. https://doi.org/10.1016/j.molcel.2017.08.003.
Mu H, Zhang T, Yang Y, Zhang D, Gao J, Li J, et al. METTL3-mediated mRNA N(6)-methyladenosine is required for oocyte and follicle development in mice. Cell Death Dis. 2021;12(11):989. https://doi.org/10.1038/s41419-021-04272-9.
Sui X, Hu Y, Ren C, Cao Q, Zhou S, Cao Y, et al. METTL3-mediated m(6)A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle. 2020;19(4):391–404. https://doi.org/10.1080/15384101.2019.1711324.
Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat Commun. 2021;12(1):1333. https://doi.org/10.1038/s41467-021-21594-6.
Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39(7):945-57.e10. https://doi.org/10.1016/j.ccell.2021.04.016.
Cheng Y, **e W, Pickering BF, Chu KL, Savino AM, Yang X, et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39(7):958-72.e8. https://doi.org/10.1016/j.ccell.2021.04.017.
Zhou T, Li S, **ang D, Liu J, Sun W, Cui X, et al. m6A RNA methylation-mediated HNF3gamma reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct Target Ther. 2020;5(1):296. https://doi.org/10.1038/s41392-020-00299-0.
Zhang Y, Gu X, Li D, Cai L, Xu Q. METTL3 regulates osteoblast differentiation and inflammatory response via smad signaling and MAPK signaling. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms21010199.
Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPbeta pathway via an m(6)A-YTHDF2-dependent manner. FASEB J. 2019;33(6):7529–44. https://doi.org/10.1096/fj.201802644R.
Tian C, Huang Y, Li Q, Feng Z, Xu Q. Mettl3 regulates osteogenic differentiation and alternative splicing of Vegfa in bone marrow mesenchymal stem cells. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20030551.
Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, et al. Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21(6):700–9. https://doi.org/10.1038/s41556-019-0318-1.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–76. https://doi.org/10.1038/nm.4416.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113(14):E2047–56. https://doi.org/10.1073/pnas.1602883113.
Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–6. https://doi.org/10.1126/science.1261417.
Ren F, Lin Q, Gong G, Du X, Dan H, Qin W, et al. Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish. Commun Biol. 2020;3(1):94. https://doi.org/10.1038/s42003-020-0827-2.
Liu HB, Muhammad T, Guo Y, Li MJ, Sha QQ, Zhang CX, et al. RNA-binding protein IGF2BP2/IMP2 is a critical maternal activator in early zygotic genome activation. Adv Sci (Weinh). 2019;6(15):1900295. https://doi.org/10.1002/advs.201900295.
Higa-Nakamine S, Suzuki T, Uechi T, Chakraborty A, Nakajima Y, Nakamura M, et al. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012;40(1):391–8. https://doi.org/10.1093/nar/gkr700.
**ng M, Liu Q, Mao C, Zeng H, Zhang X, Zhao S, et al. The 18S rRNA m(6) A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 2020;21(10):e49863. https://doi.org/10.15252/embr.201949863.
Taniguchi K, Kawai T, Kitawaki J, Tomikawa J, Nakabayashi K, Okamura K, et al. Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5’-untranslated region is related to fetal growth and preeclampsia. Faseb J. 2020;34(1):494–512. https://doi.org/10.1096/fj.201900619RR.
Bassols J, Prats-Puig A, Vázquez-Ruíz M, García-González MM, Martínez-Pascual M, Avellí P, et al. Placental FTO expression relates to fetal growth. Int J Obes (Lond). 2010;34(9):1365–70. https://doi.org/10.1038/ijo.2010.62.
Song T, Lu J, Deng Z, Xu T, Yang Y, Wei H, et al. Maternal obesity aggravates the abnormality of porcine placenta by increasing N(6)-methyladenosine. Int J Obes (Lond). 2018;42(10):1812–20. https://doi.org/10.1038/s41366-018-0113-2.
Barton SJ, Mosquera M, Cleal JK, Fuller AS, Crozier SR, Cooper C, et al. Relation of FTO gene variants to fetal growth trajectories: findings from the Southampton Women’s survey. Placenta. 2016;38:100–6. https://doi.org/10.1016/j.placenta.2015.12.015.
Kaspi A, Khurana I, Ziemann M, Connor T, Spolding B, Zimmet P, et al. Diet during pregnancy is implicated in the regulation of hypothalamic RNA methylation and risk of obesity in offspring. Mol Nutr Food Res. 2018. https://doi.org/10.1002/mnfr.201800134.
Gu Y, Chu X, Morgan JA, Lewis DF, Wang Y. Upregulation of METTL3 expression and m6A RNA methylation in placental trophoblasts in preeclampsia. Placenta. 2021;103:43–9. https://doi.org/10.1016/j.placenta.2020.10.016.
Li R, Qiu X, He M, Qiao J, He J, Zhong M. METTL3-mediated mature miR-497–5p/195–5p inhibits trophoblast migration and invasion by targeting WWP1 in preeclampsia. Cell Cycle. 2021. https://doi.org/10.1080/15384101.2021.1982527.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95. https://doi.org/10.1038/s41556-018-0045-z.
Wang J, Gao F, Zhao X, Cai Y, ** H. Integrated analysis of the transcriptome-wide m6A methylome in preeclampsia and healthy control placentas. PeerJ. 2020;8:e9880. https://doi.org/10.7717/peerj.9880.
Zhang Y, Yang H, Long Y, Zhang Y, Chen R, Shi J, et al. circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci Rep. 2021;11(1):24357. https://doi.org/10.1038/s41598-021-03662-5.
Guo Y, Song W, Yang Y. Inhibition of ALKBH5-mediated m(6) A modification of PPARG mRNA alleviates H/R-induced oxidative stress and apoptosis in placenta trophoblast. Environ Toxicol. 2022;37(4):910–24. https://doi.org/10.1002/tox.23454.
Wang J, Wang K, Liu W, Cai Y, ** H. m6A mRNA methylation regulates the development of gestational diabetes mellitus in Han Chinese women. Genomics. 2021;113(3):1048–56. https://doi.org/10.1016/j.ygeno.2021.02.016.
Qin X, Chen Y, Chen S, Liu X, Zeng W, Tian F, et al. Plasmacytoma variant translocation 1 (PVT1) regulates trophoblast viability, proliferation, and migration and is downregulated in spontaneous abortion. Am J Reprod Immunol. 2019;81(1):e13048. https://doi.org/10.1111/aji.13048.
Huppertz B. Traditional and new routes of trophoblast invasion and their implications for pregnancy diseases. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms21010289.
Huppertz B. The critical role of abnormal trophoblast development in the etiology of preeclampsia. Curr Pharm Biotechnol. 2018;19(10):771–80. https://doi.org/10.2174/1389201019666180427110547.
Xu Z, Tian P, Guo J, Mi C, Liang T, **e J, et al. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci Total Environ. 2021;776:145950. https://doi.org/10.1016/j.scitotenv.2021.145950.
Li XC, ** F, Wang BY, Yin XJ, Hong W, Tian FJ. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9(13):3853–65. https://doi.org/10.7150/thno.31868.
Qiu W, Zhou Y, Wu H, Lv X, Yang L, Ren Z, et al. RNA Demethylase FTO mediated RNA m(6)A modification is involved in maintaining maternal-fetal interface in spontaneous abortion. Front Cell Dev Biol. 2021;9:617172. https://doi.org/10.3389/fcell.2021.617172.
Chapron C, Marcellin L, Borghese B, Santulli P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat Rev Endocrinol. 2019;15(11):666–82. https://doi.org/10.1038/s41574-019-0245-z.
Vercellini P, Viganò P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–75. https://doi.org/10.1038/nrendo.2013.255.
Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–99. https://doi.org/10.1016/s0140-6736(04)17403-5.
Jiang L, Zhang M, Wu J, Wang S, Yang X, Yi M, et al. Exploring diagnostic m6A regulators in endometriosis. Aging (Albany NY). 2020;12(24):25916–38. https://doi.org/10.18632/aging.202163.
Li X, **ong W, Long X, Dai X, Peng Y, Xu Y, et al. Inhibition of METTL3/m6A/miR126 promotes the migration and invasion of endometrial stromal cells in endometriosis. Biol Reprod. 2021. https://doi.org/10.1093/biolre/ioab152.
Szubert M, Koziróg E, Olszak O, Krygier-Kurz K, Kazmierczak J, Wilczynski J. Adenomyosis and infertility—review of medical and surgical approaches. Int J Environ Res Public Health. 2021. https://doi.org/10.3390/ijerph18031235.
Bulun SE, Yildiz S, Adli M, Wei JJ. Adenomyosis pathogenesis: insights from next-generation sequencing. Hum Reprod Update. 2021;27(6):1086–97. https://doi.org/10.1093/humupd/dmab017.
Cunningham RK, Horrow MM, Smith RJ, Springer J. Adenomyosis: a sonographic diagnosis. Radiographics. 2018;38(5):1576–89. https://doi.org/10.1148/rg.2018180080.
Bird CC, McElin TW, Manalo-Estrella P. The elusive adenomyosis of the uterus—revisited. Am J Obstet Gynecol. 1972;112(5):583–93. https://doi.org/10.1016/0002-9378(72)90781-8.
Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen ZJ, et al. m(6)A RNA methylation regulators contribute to eutopic endometrium and myometrium dysfunction in adenomyosis. Front Genet. 2020;11:716. https://doi.org/10.3389/fgene.2020.00716.
Wang Q, Wei Y, Zhang J. Combined knockdown of D-dopachrome tautomerase and migration inhibitory factor inhibits the proliferation, migration, and invasion in human cervical cancer. Int J Gynecol Cancer. 2017;27(4):634. https://doi.org/10.1097/IGC.0000000000000951.
Zhang S, Deng W, Liu Q, Wang P, Yang W, Ni W. Altered m(6) A modification is involved in up-regulated expression of FOXO3 in luteinized granulosa cells of non-obese polycystic ovary syndrome patients. J Cell Mol Med. 2020;24(20):11874–82. https://doi.org/10.1111/jcmm.15807.
Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. Embo J. 2020;39(12):e103181. https://doi.org/10.15252/embj.2019103181.
Zhang C, Hu J, Wang W, Sun Y, Sun K. HMGB1-induced aberrant autophagy contributes to insulin resistance in granulosa cells in PCOS. FASEB J. 2020;34(7):9563–74. https://doi.org/10.1096/fj.202000605RR.
Zhou L, Han X, Li W, Wang N, Yao L, Zhao Y, et al. N6-methyladenosine Demethylase FTO induces the dysfunctions of ovarian granulosa cells by upregulating Flotillin 2. Reprod Sci. 2021. https://doi.org/10.1007/s43032-021-00664-6.
Zou J, Li Y, Liao N, Liu J, Zhang Q, Luo M, et al. Identification of key genes associated with polycystic ovary syndrome (PCOS) and ovarian cancer using an integrated bioinformatics analysis. J Ovarian Res. 2022;15(1):30. https://doi.org/10.1186/s13048-022-00962-w.
Brodie MJ, Mintzer S, Pack AM, Gidal BE, Vecht CJ, Schmidt D. Enzyme induction with antiepileptic drugs: cause for concern? Epilepsia. 2013;54(1):11–27. https://doi.org/10.1111/j.1528-1167.2012.03671.x.
Morgan S, Anderson RA, Gourley C, Wallace WH, Spears N. How do chemotherapeutic agents damage the ovary? Hum Reprod Update. 2012;18(5):525–35. https://doi.org/10.1093/humupd/dms022.
Blumenfeld Z. Preservation of fertility and ovarian function and minimalization of chemotherapy associated gonadotoxicity and premature ovarian failure: the role of inhibin-A and -B as markers. Mol Cell Endocrinol. 2002;187(1–2):93–105. https://doi.org/10.1016/s0303-7207(01)00712-2.
Conway GS. Premature ovarian failure. Br Med Bull. 2000;56(3):643–9. https://doi.org/10.1258/0007142001903445.
Huang B, Ding C, Zou Q, Wang W, Li H. Cyclophosphamide regulates N6-methyladenosine and m6A RNA enzyme levels in human granulosa cells and in ovaries of a premature ovarian aging mouse model. Front Endocrinol (Lausanne). 2019;10:415. https://doi.org/10.3389/fendo.2019.00415.
Ding C, Zou Q, Ding J, Ling M, Wang W, Li H, et al. Increased N6-methyladenosine causes infertility is associated with FTO expression. J Cell Physiol. 2018;233(9):7055–66. https://doi.org/10.1002/jcp.26507.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. https://doi.org/10.1016/j.molcel.2012.10.015.
Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N(6)-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020;11(10):911. https://doi.org/10.1038/s41419-020-03071-y.
Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11(1):2578. https://doi.org/10.1038/s41467-020-16306-5.
Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10(1):2300. https://doi.org/10.1038/s41467-019-10246-5.
Lin X, Wang F, Chen J, Liu J, Lin YB, Li L, et al. N(6)-methyladenosine modification of CENPK mRNA by ZC3H13 promotes cervical cancer stemness and chemoresistance. Mil Med Res. 2022;9(1):19. https://doi.org/10.1186/s40779-022-00378-z.
Liang L, Zhu Y, Li J, Zeng J, Wu L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res. 2022;41(1):261. https://doi.org/10.1186/s13046-022-02462-7.
Zhou S, Bai ZL, **a D, Zhao ZJ, Zhao R, Wang YY, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57(5):590–7. https://doi.org/10.1002/mc.22782.
Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/beta-catenin signalling pathway. Cell Prolif. 2020;53(3):e12768. https://doi.org/10.1111/cpr.12768.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–83. https://doi.org/10.1038/s41556-018-0174-4.
Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021;11(3):1100–14. https://doi.org/10.7150/thno.49345.
Ruan P, Wang S, Yang C, Huang X, Sun P, Tan A. m(6)A mRNA methylation regulates the ERK/NF-κB/AKT signaling pathway through the PAPPA/IGFBP4 axis to promote proliferation and tumor formation in endometrial cancer. Cell Biol Toxicol. 2022. https://doi.org/10.1007/s10565-022-09751-z.
Xue T, Liu X, Zhang M, Qiukai E, Liu S, Zou M, et al. PADI2-catalyzed MEK1 citrullination activates ERK1/2 and promotes IGF2BP1-mediated SOX2 mRNA stability in endometrial cancer. Adv Sci (Weinh). 2021;8(6):2002831. https://doi.org/10.1002/advs.202002831.
Zhang C, Liu J, Guo H, Hong D, Ji J, Zhang Q, et al. m6A RNA methylation regulators were associated with the malignancy and prognosis of ovarian cancer. Bioengineered. 2021;12(1):3159–76. https://doi.org/10.1080/21655979.2021.1946305.
Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, et al. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20(1):45. https://doi.org/10.1186/s12943-021-01340-8.
Liu T, Wei Q, ** J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816–31. https://doi.org/10.1093/nar/gkaa048.
Müller S, Glaß M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47(1):375–90. https://doi.org/10.1093/nar/gky1012.
Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, et al. FTO-dependent N (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 2020;80(16):3200–14. https://doi.org/10.1158/0008-5472.Can-19-4044.
Sun M, Zhang X, Bi F, Wang D, Zhou X, Li X, et al. FTO inhibits epithelial ovarian cancer progression by destabilising SNAI1 mRNA through IGF2BP2. Cancers (Basel). 2022. https://doi.org/10.3390/cancers14215218.
Sun R, Yuan L, Jiang Y, Wan Y, Ma X, Yang J, et al. ALKBH5 activates FAK signaling through m6A demethylation in ITGB1 mRNA and enhances tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer. Theranostics. 2023;13(2):833–48. https://doi.org/10.7150/thno.77441.
Zhang Y, Qiu JG, Jia XY, Ke Y, Zhang MK, Stieg D, et al. METTL3-mediated N6-methyladenosine modification and HDAC5/YY1 promote IFFO1 downregulation in tumor development and chemo-resistance. Cancer Lett. 2023;553:215971. https://doi.org/10.1016/j.canlet.2022.215971.
Yu HL, Ma XD, Tong JF, Li JQ, Guan XJ, Yang JH. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther. 2019;12:6191–201. https://doi.org/10.2147/OTT.S205730.
Ma Z, Li Q, Liu P, Dong W, Zuo Y. METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP. Cell Biol Int. 2020;44(12):2524–31. https://doi.org/10.1002/cbin.11459.
Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. https://doi.org/10.1038/nrm.2016.132.
Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22(2):119–31. https://doi.org/10.1038/s41576-020-00295-8.
Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20(1):18. https://doi.org/10.1186/s12943-020-01263-w.
Ma J, Song B, Wei Z, Huang D, Zhang Y, Su J, et al. m5C-Atlas: a comprehensive database for decoding and annotating the 5-methylcytosine (m5C) epitranscriptome. Nucleic Acids Res. 2022;50(D1):D196-d203. https://doi.org/10.1093/nar/gkab1075.
Ding C, Lu J, Li J, Hu X, Liu Z, Su H, et al. RNA-methyltransferase Nsun5 controls the maternal-to-zygotic transition by regulating maternal mRNA stability. Clin Transl Med. 2022;12(12):e1137. https://doi.org/10.1002/ctm2.1137.
Gu WX, Chen Y, Wang W. Immune infiltrates of m5C RNA methylation-related LncRNAs in uterine corpus endometrial carcinoma. J Oncol. 2022;2022:1531474. https://doi.org/10.1155/2022/1531474.
Dong C, Dang L, Gao X, Xu R, Zhang H, Zhang X. Systematic analysis of tumor microenvironment patterns and oxidative stress characteristics of endometrial carcinoma mediated by 5-methylcytosine regulators. Oxid Med Cell Longev. 2022;2022:6431164. https://doi.org/10.1155/2022/6431164.
Wang L, Gao S. Identification of 5-methylcytosine-related signature for predicting prognosis in ovarian cancer. Biol Res. 2021;54(1):18. https://doi.org/10.1186/s40659-021-00340-8.
Sha C, Chen L, Lin L, Li T, Wei H, Yang M, et al. TRDMT1 participates in the DNA damage repair of granulosa cells in premature ovarian failure. Aging (Albany NY). 2021;13(11):15193–213. https://doi.org/10.18632/aging.203080.
Zou F, Tu R, Duan B, Yang Z, ** Z, Song X, et al. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc Natl Acad Sci USA. 2020;117(7):3603–9. https://doi.org/10.1073/pnas.1910862117.
Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L, et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther. 2022;7(1):334. https://doi.org/10.1038/s41392-022-01175-9.
Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551(7679):251–5. https://doi.org/10.1038/nature24456.
Luo Y, Yao Y, Wu P, Zi X, Sun N, He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J Hematol Oncol. 2022;15(1):63. https://doi.org/10.1186/s13045-022-01285-5.
Liu J, Geng R, Zhong Z, Zhang Y, Ni S, Liu W, et al. N1-Methyladenosine-related lncRNAs are potential biomarkers for predicting prognosis and immune response in uterine corpus endometrial carcinoma. Oxid Med Cell Longev. 2022;2022:2754836. https://doi.org/10.1155/2022/2754836.
Zhao J, Zou J, Jiao W, Lin L, Wang J, Lin Z. Construction of N-7 methylguanine-related mRNA prognostic model in uterine corpus endometrial carcinoma based on multi-omics data and immune-related analysis. Sci Rep. 2022;12(1):18813. https://doi.org/10.1038/s41598-022-22879-6.
Sun J, Li L, Chen H, Gan L, Guo X, Sun J. Identification and validation of an m7G-related lncRNAs signature for prognostic prediction and immune function analysis in endometrial cancer. Genes (Basel). 2022. https://doi.org/10.3390/genes13081301.
Chen R, Sun K, Hou Y, Shen J, Chen J, Dong F, et al. Identification of m7G methylation-related miRNA signature associated with survival and immune microenvironment regulation in uterine corpus endometrial carcinoma. Biomed Res Int. 2022;2022:8776678. https://doi.org/10.1155/2022/8776678.
Ge J, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci. 2013;38(4):210–8. https://doi.org/10.1016/j.tibs.2013.01.002.
Li H, Chen L, Han Y, Zhang F, Wang Y, Han Y, et al. The identification of RNA modification gene PUS7 as a potential biomarker of ovarian cancer. Biology (Basel). 2021. https://doi.org/10.3390/biology10111130.
Liu X, **ao M, Zhang L, Li L, Zhu G, Shen E, et al. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021;35(3):e23655. https://doi.org/10.1002/jcla.23655.
** D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2021;14(1):32. https://doi.org/10.1186/s13045-021-01048-8.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677-91.e10. https://doi.org/10.1016/j.ccell.2019.03.006.
Liu WW, Wang H, Zhu XY. Physio-pathological effects of N6-methyladenosine and its therapeutic implications in leukemia. Biomark Res. 2022;10(1):64. https://doi.org/10.1186/s40364-022-00410-3.
Zheng X, Gong Y. Functions of RNA N(6)-methyladenosine modification in acute myeloid leukemia. Biomark Res. 2021;9(1):36. https://doi.org/10.1186/s40364-021-00293-w.
Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020;10(8):2546–54.
Zheng F, Du F, Zhao J, Wang X, Si Y, ** P, et al. The emerging role of RNA N6-methyladenosine methylation in breast cancer. Biomark Res. 2021;9(1):39. https://doi.org/10.1186/s40364-021-00295-8.
Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. https://doi.org/10.1186/s12943-020-01170-0.
Yan J, Huang X, Zhang X, Chen Z, Ye C, **ang W, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521(4):887–93. https://doi.org/10.1016/j.bbrc.2019.11.016.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193–205. https://doi.org/10.1136/gutjnl-2019-319639.
Ji H, Zhang JA, Liu H, Li K, Wang ZW, Zhu X. Comprehensive characterization of tumor microenvironment and m6A RNA methylation regulators and its effects on PD-L1 and immune infiltrates in cervical cancer. Front Immunol. 2022;13:976107. https://doi.org/10.3389/fimmu.2022.976107.
Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N(6)-Methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019;79(11):2812–20. https://doi.org/10.1158/0008-5472.Can-18-3592.
Liu K, Ouyang QY, Zhan Y, Yin H, Liu BX, Tan LM, et al. Pharmacoepitranscriptomic landscape revealing m6A modification could be a drug-effect biomarker for cancer treatment. Mol Ther Nucleic Acids. 2022;28:464–76. https://doi.org/10.1016/j.omtn.2022.04.001.
Nie S, Zhang L, Liu J, Wan Y, Jiang Y, Yang J, et al. ALKBH5-HOXA10 loop-mediated JAK2 m6A demethylation and cisplatin resistance in epithelial ovarian cancer. J Exp Clin Cancer Res. 2021;40(1):284. https://doi.org/10.1186/s13046-021-02088-1.
Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y, et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m(6)A-MYC expression. Int J Biol Sci. 2022;18(2):507–21. https://doi.org/10.7150/ijbs.67770.
Funding
This work was supported by grant from National Natural Science Foundation of China (No. 81000257).
Author information
Authors and Affiliations
Contributions
LC proposed the idea and reviewed the manuscript, EH drafted and revised the initial manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huang, E., Chen, L. RNA N6-methyladenosine modification in female reproductive biology and pathophysiology. Cell Commun Signal 21, 53 (2023). https://doi.org/10.1186/s12964-023-01078-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12964-023-01078-4