Abstract
Background
Lipid droplet (LD)-laden microglia is a key pathological hallmark of multiple sclerosis. The recent discovery of this novel microglial subtype, lipid-droplet-accumulating microglia (LDAM), is notable for increased inflammatory factor secretion and diminished phagocytic capability. Lipophagy, the autophagy-mediated selective degradation of LDs, plays a critical role in this context. This study investigated the involvement of microRNAs (miRNAs) in lipophagy during demyelinating diseases, assessed their capacity to modulate LDAM subtypes, and elucidated the potential underlying mechanisms involved.
Methods
C57BL/6 mice were used for in vivo experiments. Two weeks post demyelination induction at cervical level 4 (C4), histological assessments and confocal imaging were performed to examine LD accumulation in microglia within the lesion site. Autophagic changes were observed using transmission electron microscopy. miRNA and mRNA multi-omics analyses identified differentially expressed miRNAs and mRNAs under demyelinating conditions and the related autophagy target genes. The role of miR-223 in lipophagy under these conditions was specifically explored. In vitro studies, including miR-223 upregulation in BV2 cells via lentiviral infection, validated the bioinformatics findings. Immunofluorescence staining was used to measure LD accumulation, autophagy levels, target gene expression, and inflammatory mediator levels to elucidate the mechanisms of action of miR-223 in LDAM.
Results
Oil Red O staining and confocal imaging revealed substantial LD accumulation in the demyelinated spinal cord. Transmission electron microscopy revealed increased numbers of autophagic vacuoles at the injury site. Multi-omics analysis revealed miR-223 as a crucial regulatory gene in lipophagy during demyelination. It was identified that cathepsin B (CTSB) targets miR-223 in autophagy to integrate miRNA, mRNA, and autophagy gene databases. In vitro, miR-223 upregulation suppressed CTSB expression in BV2 cells, augmented autophagy, alleviated LD accumulation, and decreased the expression of the inflammatory mediator IL-1β.
Conclusion
These findings indicate that miR-223 plays a pivotal role in lipophagy under demyelinating conditions. By inhibiting CTSB, miR-223 promotes selective LD degradation, thereby reducing the lipid burden and inflammatory phenotype in LDAM. This study broadens the understanding of the molecular mechanisms of lipophagy and proposes lipophagy induction as a potential therapeutic approach to mitigate inflammatory responses in demyelinating diseases.
Similar content being viewed by others
Introduction
Multiple sclerosis (MS) is a demyelinating disease characterized by central nervous system inflammation, resulting in impaired nerve impulse transmission due to loss of the axonal myelin sheath, accompanied by neurodegeneration [1]. In MS, myelin phospholipids and neuronal fragments cause neuroinflammation, further exacerbating demyelination [2, 3]. Microglia, as resident phagocytes of the central nervous system, play a fundamental role in the clearance of myelin debris [4]. Microglia facilitate the remyelination process by providing nutritional support to oligodendrocytes [4]. However, in MS, the capacity of microglia for clearance is limited [5]. The continuous internalization of myelin debris overburdens microglia, leading to the accumulation of numerous LDs within these cells. These LD-rich microglia, also known as lipid-droplet-accumulating microglia (LDAM), exhibit unique transcriptional characteristics associated with cellular dysfunction. These cells exhibit significant phagocytic defects, and increased lipid storage. These LDAM activities hinder remyelination, resulting in neurofunctional impairment [6,7,8]. Modulating LDAM can thus prevent inflammatory lipid distribution and improve neural function [9].
Autophagy within microglia regulates lipid homeostasis. However, impaired autophagy results in LD accumulation and metabolic dysfunction [5A). Of particular interest was Ctsb, which is known for its dual roles as a lysosomal cathepsin and a regulator of autophagy and closely linked to the metabolism of lipids. This finding, coupled with the observed lysosomal alterations induced by miR-223 in BV2 cells, led us to hypothesize that Ctsb may serve as a critical target for miR-223 in the regulation of lipophagy.
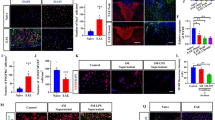
Identification of Ctsb as a direct target of miR-223-5p. (A) Prediction of autophagy-related miR-223 target gene candidates. (B) The q-PCR results showed that miR-223 regulates Ctsb expression at the posttranscriptional level. (C) The predicted target sequence of miR-223-5p in the 3′-UTR of Ctsb. (D) Luciferase activity in miR-223 NC and miR-223 OE cells transfected with the Ctsb-3UTR-wt or Ctsb-3UTR-mut reporter plasmid. Control: untreated BV2 cells; LPS: BV2 cells treated with LPS; miR-223 NC + LPS: miR-223 NC cells treated with LPS; miR-223 OE + LP: miR-223 OE cells treated with LPS. All the data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Subsequent investigations aimed to determine whether miR-223 regulates Ctsb expression at the post-transcriptional level. The results confirmed this hypothesis: Overexpression of miR-223 significantly repressed Ctsb mRNA expression (control: 1.00 ± 0.008, LPS: 3.20 ± 0.005, miR-223 NC + LPS: 3.36 ± 0.192, and miR-223 OE + LPS: 1.527 ± 0.033 (mean ± SEM) (Fig. 5B). To further determine whether miR-223 downregulates Ctsb expression through binding to seed sequences, TargetScan was used to predict potential miR-223 binding sites within the 3′-UTR of Ctsb (Fig. 5C). Using the wild-type Ctsb reporter construct, luciferase activity was significantly decreased upon miR-223 overexpression as compared to the mutated construct (Fig. 5D). By reducing luciferase activity, miR-223 was demonstrated to directly interact with Ctsb mRNA, substantiating its role in post-transcriptional regulation.
6. MiR-223 inhibits CTSB and promotes lipophagy
Then the impact of miR-223 was explored on CTSB and lipophagy in BV2 cells. Following LPS stimulation, CTSB expression was assessed using immunofluorescence confocal microscopy. The results showed significantly lower CTSB expression in the miR-223 OE group than in the BV2 and miR-223 NC groups after LPS stimulation (1 ± 0.038 in the control group, 3.720 ± 0.182 in the LPS group, 3.639 ± 0.219 in the miR-223 NC + LPS group, and 2.459 ± 0.174 in the miR-223 OE + LPS group (mean ± SEM)) (Fig. 6A-C). Western blot analysis confirmed this trend (Fig. 6D, E). These results demonstrated the inhibitory effect of miR-223 on CTSB expression.
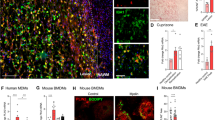
MiR-223 inhibits CTSB and promotes lipophagy. (A) Representative immunofluorescence images showing that the upregulation of miR-223 inhibited CTSB expression in BV2 cells (scale bar = 10 μm). (B) Partial magnification of Figure A (scale bar = 2 μm). (C) Quantitative CTSB fluorescence analysis showed that CTSB density in BV2 cells was significantly decreased after miR-223 upregulation. (D, E) Western blotting images and statistical analysis showed that miR-223 decreased the protein level of CTSB. (F, G) Immunofluorescence staining of spinal cord samples showing BODIPY + LDs (yellow), LC3 (pink), and DAPI (blue). (H) Quantification of LC3 + densities showed a significant increase in LC3 + densities in the miR-223 OE + LPS group. (I) BODIPY + density quantification showed that BODIPY + density decreased significantly in the miR-223 OE + LPS group. Control: untreated BV2 cells; LPS: BV2 cells treated with LPS; miR-223 NC + LPS: miR-223 NC cells treated with LPS; miR-223 OE + LP: miR-223 OE cells treated with LPS. All the data are presented as the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001
Furthermore, using LC3 and BODIPY fluorescence double staining, it was investigated whether miR-223 regulates selective autophagy in BV2 cells and its effect on LDs. Confocal imaging after LPS stimulation showed enhanced LC3 and BODIPY colocalization in BV2 cells, indicating increased lipophagy, with the miR-223 OE group showing an even greater effect (Fig. 6F, G). Statistical analysis revealed increased LC3 fluorescence intensity in the miR-223 OE + LPS group (0.96 ± 0.071 in the control group, 3.38 ± 0.469 in the LPS group, 3.35 ± 0.498 in the miR-223 NC + LPS group, and 5.19 ± 0.532 in the miR-223 OE + LPS group (mean ± SEM) (Fig. 6H) and a significant reduction in BODIPY fluorescence intensity in the miR-223 OE group (1.00 ± 0.045 in the control group, 2.95 ± 0.378 in the LPS group, 3.37 ± 0.394 in the miR-223 NC + LPS group, and 1.78 ± 0.230 in the miR-223 OE + LPS group (mean ± SEM)) (Fig. 6I). LC3 and BODIPY fluorescence intensities showed a negative correlation. These results highlight the role of miR-223 in enhancing LDAM lipophagy and accelerating intracellular LD degradation.
7. MiR-223 inhibits the expression of the inflammatory factor IL-1β
IL-1β is synthesized and secreted by a diverse array of immune and nonimmune cells upon exposure to inflammatory stimuli, and its expression is rapidly upregulated following LPS induction. Consequently, IL-1β concentrations serve as a barometer for the inflammatory state of immune cells. To elucidate the influence of miR-223 on the microglial inflammatory response, IL-1β expression levels across various experimental groups were assessed utilizing both immunofluorescence staining and q-PCR methodologies. Confocal imaging revealed increased IL-1β expression in control BV2 and miR-223 NC cells after LPS stimulation, while miR-223-overexpressing microglia exhibited decreased IL-1β levels (Fig. 7A). Statistical analyses provided robust evidence for the suppression of IL-1β following the overexpression of miR-223, with the measured values indicating a notable decrease (control group: 1 ± 0.1114, LPS group: 3.425 ± 0.6000, miR-223 NC + LPS group: 3.168 ± 0.4519, and miR-223 OE + LPS group: 1.515 ± 0.1619 (mean ± SEM)) (Fig. 7B). Similarly, q-PCR results mirrored this pattern, revealing a significant reduction in IL-1β expression (control group: 1.02 ± 0.130, LPS group: 5.46 ± 0.570, miR-223 NC + LPS group: 4.94 ± 0.16, and miR-223 OE + LPS group: 2.42 ± 0.496 (mean ± SEM)) (Fig. 7C). These findings collectively underscore the inhibitory impact of miR-223 overexpression on IL-1β production, consistent with the hypothesis that miR-223 plays a critical role in modulating the microglial inflammatory response. In summary, miR-223 appears to promote LD degradation by inhibiting CTSB, thereby enhancing lipophagy in LDAM and suppressing cellular inflammatory responses.
MiR-223 inhibits the expression of the inflammatory factor IL-1β. (A, B) Immunofluorescence images showing that the expression of the inflammatory factor IL-1β decreased after upregulation of miR-223. (C) q-PCR results showing that miR-223 decreased the mRNA level of IL-1β. Control: untreated BV2 cells; LPS: BV2 cells treated with LPS; miR-223 NC + LPS: miR-223 NC cells treated with LPS; miR-223 OE + LP: miR-223 OE cells treated with LPS. All the data are presented as the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001
Discussion
In this study, LDAM, a pathological cell subtype characterized by phagocytic defects and excessive LDs, was identified as prevalent in demyelinating lesions. Through a comprehensive multiomics analysis of miRNAs and mRNAs in demyelinated mouse spinal cord tissue, miR-223 was experimentally demonstrated as a novel regulator of lipophagy. The upregulation of miR-223 enhanced LPC-induced autophagy, which was coupled with a decrease in the number of LDs. Furthermore, CTSB was predicted as its target, and the experimental results revealed that the upregulation of miR-223 did inhibit the expression of CTSB and IL-1β in BV2 cells. In conclusion, it was demonstrated that the upregulation of miR-223 inhibits CTSB, promotes lipophagy-mediated LD degradation, and reduces the inflammatory response in microglia.
In recent years, the development of bioinformatics has accelerated progress in the study of human disease mechanisms. GO analysis revealed several terms related to demyelinating diseases, including regulation of immune response, microglial activation, and lipid metabolism. Microglia, the resident macrophages of the CNS [26], play a crucial role in clearing myelin debris from demyelinated sites for efficient remyelination and disease progression attenuation [27]. Initially, myelin-laden microglia exhibit a protective phenotype, characterized by the release of immunosuppressive and repair factors [28,29,30]. However, prolonged accumulation of myelin-derived lipids leads to the formation of numerous LDs, shifting microglia toward a phenotype that inhibits remyelination [5]. By inducing demyelinating injury in the mouse spinal cord, it was observed that the activation of numerous microglia laden with abundant LDs. The expression of ASC, which can regulate the activation of inflammasomes [31], was found to be high in LD-rich microglia at the site of demyelination, suggesting an obvious proinflammatory state. Other research groups have also identified LD-rich microglia in an aging mouse model. This subtype of microglia has phagocytic defects and secretes excessive proinflammatory cytokines, altering the immune microenvironment into a more inflammatory state [6]. Hence, this new subtype was named LDAM [6]. LD-rich microglia exhibit significantly reduced phagocytosis of myelin debris, indicating an association between increased damaged lipid storage and phagocytic defects [25]. Therefore, promoting LD degradation in LDAM at injury sites is crucial for restoring the phagocytic function and normal phenotype of microglia in treating demyelinating diseases.
Transcriptomic bioinformatics analyses have shown that autophagy is a key factor in demyelinating diseases. Histological staining of demyelinated spinal cord tissue revealed the presence of LDAM, along with enhanced autophagy in microglia at injury sites. Changes in microglial autophagy can regulate immune responses and neuroprotective effects [32,33,34], and prevent cellular senescence [35]. Enhanced autophagy has also been shown to reduce LD accumulation in foam cells [17, 43,44,45,46]. MiR-223 has been shown to regulate cholesterol metabolism and inflammatory signaling pathways by targeting cholesterol biosynthesis pathways, reversing foam cell formation in VSMC macrophages [47]. It can also activate the PI3K/AKT pathway to block TLR4 signaling and inhibit atherosclerosis development [48]. In the nervous system, miR-223 plays a crucial role in promoting the effective activation and phagocytosis of myelin debris by microglia, which are essential for initiating remyelination [49]. It facilitates microglial differentiation into an efficient M2 phenotype with enhanced phagocytic activity, inhibits NF-κB and the NLRP3 inflammasome, and regulates the immune microenvironment at lesion sites [47, 50, 51], aiding in repair while suppressing neuroinflammation. This discovery holds significant pathological and physiological relevance for multiple sclerosis and other neurodegenerative diseases. Nevertheless, the function of miR-223 in modulating lipophagy within the nervous system remains undocumented.
In vitro, LPS can induce BV2 cells to adopt the LDAM phenotype. A significant increase in LC3 expression and lysosome numbers was observed in LDAM. This phenomenon may be due to the following reason: during demyelination or LPS-induced acute inflammation, the autophagy-lysosome system in microglia is burdened with lipid digestion, leading to a lysosomal storage phenotype and pushing degradation and metabolism mechanisms to their limits. If lipid overload cannot be compensated by lysosomal digestion, substantial LD accumulation and the transformation of microglia to the LDAM subtype result. Hence, enhancing selective autophagy targeting LDs appears to be a feasible strategy for reversing the LDAM subtype. Fortunately, miR-223 can promote selective degradation of LDs by autophagy: the upregulation of miR-223 in LPS-stimulated BV2 cells enhanced intracellular autophagy levels and inhibited LD accumulation and IL-1β expression, thereby improving LDAM subtype transformation. It was indicated that miR-223 is an important regulatory factor in autophagy and lipid metabolism in other disease models [52,53,54] and the serum samples of ASO patients [54]. Overexpression of miR-223 in the VSMCs of ASO patients induces autophagy, significantly inhibiting foam cell formation and reducing intracellular total cholesterol levels [54]. Blocking autophagy with Atg7 siRNA was shown to weaken the inhibitory effect of miR-223 overexpression on foam cell formation, suggesting that miR-223 overexpression partially inhibits foam cell formation in VSMCs by inducing autophagy [54]. In this study, it was confirmed that miR-223 promotes lipid metabolism and inhibits the transformation of microglia into the LDAM subtype, improving the inflammatory response through lipophagy in microglia.
To identify the downstream target genes of miR-223 in lipophagy during demyelination, the miRNA, mRNA, and autophagy gene databases were analyzed collectively. The result showed that CTSB was a key target gene regulated by miR-223 in lipophagy. Previous studies have shown that cysteine proteases are crucial for protein hydrolysis and degradation in both lysosomal and extralysosomal environments and play indispensable roles in autophagy, antigen presentation, cellular stress signaling, metabolism, and lysosome-dependent cell death [55,56,57]. Whole-genome expression analysis has revealed that bone marrow-derived macrophages (BMDMs) lacking CTSB exhibit increased expression of TFEB, a central transcription factor that controls lysosomal and autophagy-related gene expression [26]. Transmission electron microscopy has revealed increased numbers and sizes of lysosomes and autophagosomes in CTSB-deficient BMDMs [56]. These results suggest that CTSB may inhibit the autophagy‒lysosome process under steady-state conditions. CTSB homeostasis is also critical for traumatic brain injury repair [58]. Bioinformatics analysis indicated that CTSB gene expression was upregulated in a demyelinated state, and a similar trend was observed in BV2 cells after LPS stimulation. It could be interpreted that the organism achieves relative homeostasis through feedback regulation., with negative feedback being more pronounced. Enhanced autophagy during demyelination or after LPS stimulation of microglia leads to upregulation of the negative regulatory factor CTSB to achieve negative feedback regulation of autophagy. Interestingly, miR-223 upregulation inhibited Ctsb, thereby promoting lipophagy.
Strengths and limitations of the study
This study has several strengths. MiRNA and mRNA multi-omics analyses were utilized to determine the close relationship between miR-223 and lipophagy following demyelination, and Ctsb was identified as a potential target of miR-223 through database analysis. This is the advantage of the methodology. There are few studies on miRNAs related to lipophagy after demyelination, but the experimental results proved that the upregulation of miR-223 can promote lipophagy in microglia, which fills a theoretical gap in this area.
This study also has important limitations. First, due to the small size of LPC lesions, some intact tissue was present in the samples, potentially leading to a higher false-negative rate of differential genes and overlooking other key genes. Second, it was validated the miR-223-related pathway only in cellular experiments, which cannot fully simulate the complex physiological environment or directly reflect intricate interactions within organisms. Fortunately, other teams have demonstrated through in vivo experiments that miR-223 promotes the effective activation and phagocytosis of myelin debris by microglia [49]. Finally, the experimental design lacked a miR-223 knockdown or downregulated group. The aim was to study therapeutic factors post-demyelination, it was demonstrated that miR-223 has a positive effect in promoting autophagy and LD degradation; however, the opposite trend in microglia lacking miR-223 was not validated in this study.
Conclusions
Microglia transformed into LDAM after inflammation-induced spinal cord demyelination. Overexpression of miR-223 effectively inhibited the inhibitory effect of CTSB on microglial autophagy, reducing LD accumulation and the release of inflammatory mediators. Thus, this study provides a new therapeutic idea for demyelinating diseases: inhibiting the inflammatory microenvironment by promoting lipophagy to suppress lipid droplets.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Alizadeh A, Dyck SM, Karimi-Abdolrezaee S. Myelin damage and repair in pathologic CNS: challenges and prospects. Front Mol Neurosci. 2015;8:35.
Lassmann H, van Horssen J, Mahad D. Progressive multiple sclerosis: pathology and pathogenesis. Nat Rev Neurol. 2012;8(11):647–56.
Yao F, Luo Y, Chen Y, Li Y, Hu X, You X, Li Z, Yu S, Tian D, Zheng M, et al. Myelin debris impairs tight junctions and promotes the Migration of Microvascular endothelial cells in the injured spinal cord. Cell Mol Neurobiol. 2023;43(2):741–56.
Olah M, Amor S, Brouwer N, Vinet J, Eggen B, Biber K, Boddeke HW. Identification of a microglia phenotype supportive of remyelination. GLIA. 2012;60(2):306–21.
Bogie J, Grajchen E, Wouters E, Corrales AG, Dierckx T, Vanherle S, Mailleux J, Gervois P, Wolfs E, Dehairs J et al. Stearoyl-CoA desaturase-1 impairs the reparative properties of macrophages and microglia in the brain. J Exp Med 2020, 217(5).
Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci. 2020;23(2):194–208.
Miron VE, Boyd A, Zhao JW, Yuen TJ, Ruckh JM, Shadrach JL, van Wijngaarden P, Wagers AJ, Williams A, Franklin R, et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci. 2013;16(9):1211–8.
Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Bio. 2019;20(3):137–55.
Loving BA, Tang M, Neal MC, Gorkhali S, Murphy R, Eckel RH, Bruce KD. Lipoprotein lipase regulates microglial lipid droplet accumulation. Cells Basel. 2021;10(2).
Xu Y, Propson NE, Du S, **ong W, Zheng H. Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. P Natl Acad Sci USA. 2021;118(27).
Singh R, Kaushik S, Wang Y, **ang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5.
Chung J, Park J, Lai ZW, Lambert TJ, Richards RC, Zhang J, Walther TC, Farese RJ. The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets. Nat Cell Biol. 2023;25(8):1101–10.
Laval T, Ouimet M. A role for lipophagy in atherosclerosis. Nat Rev Cardiol. 2023;20(7):431–2.
Pu M, Zheng W, Zhang H, Wan W, Peng C, Chen X, Liu X, Xu Z, Zhou T, Sun Q, et al. ORP8 acts as a lipophagy receptor to mediate lipid droplet turnover. Protein Cell. 2023;14(9):653–67.
Wang Z, Zhang H. Join the club: ORP8 is a lipophagy receptor. Protein Cell. 2023;14(9):632–4.
Haidar M, Loix M, Vanherle S, Dierckx T, Vangansewinkel T, Gervois P, Wolfs E, Lambrichts I, Bogie J, Hendriks J. Targeting lipophagy in macrophages improves repair in multiple sclerosis. Autophagy. 2022;18(11):2697–710.
Jeffries J, Zhou W, Hsu AY, Deng Q. miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. 2019;451:136–41.
Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–6.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34.
Yang Y, Liang C. MicroRNAs: an emerging player in autophagy. ScienceOpen Res. 2015;2015.
Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in Cancer. Curr Genomics 2010;11(7):537–61.
Lee DH, Park SH, Ahn J, Hong SP, Lee E, Jang YJ, Ha TY, Huh YH, Ha SY, Jeon TI, et al. Mir214-3p and Hnf4a/Hnf4α reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy. 2021;17(9):2415–31.
Shao W, Wang S, Wang X, Yao L, Yuan X, Huang D, Lv B, Ye Y, Xue H. miRNA-29a inhibits atherosclerotic plaque formation by mediating macrophage autophagy via PI3K/AKT/mTOR pathway. Aging. 2022;14(5):2418–31.
Ma J, Yang S, Ma A, Pan X, Wang H, Li N, Liu S, Wu M. Expression of miRNA-155 in carotid atherosclerotic plaques of apolipoprotein E knockout (ApoE(-/-)) mice and the interventional effect of rapamycin. Int Immunopharmacol. 2017;46:70–4.
Zhou LQ, Dong MH, Hu ZW, Tang Y, Chu YH, Chen M, Yang S, Chen Z, Wu LJ, Wang W, et al. Staged suppression of microglial autophagy facilitates regeneration in CNS demyelination by enhancing the production of linoleic acid. P Natl Acad Sci USA. 2023;120(1):e2084977176.
Devanney NA, Stewart AN, Gensel JC. Microglia and macrophage metabolism in CNS injury and disease: the role of immunometabolism in neurodegeneration and neurotrauma. Exp Neurol. 2020;329:113310.
Sen MK, Mahns DA, Coorssen JR, Shortland PJ. The roles of microglia and astrocytes in phagocytosis and myelination: insights from the cuprizone model of multiple sclerosis. GLIA. 2022;70(7):1215–50.
Bogie JF, Timmermans S, Huynh-Thu VA, Irrthum A, Smeets HJ, Gustafsson JÅ, Steffensen KR, Mulder M, Stinissen P, Hellings N, et al. Myelin-derived lipids modulate macrophage activity by liver X receptor activation. PLoS One. 2012;7(9):e44998.
Bogie JF, Jorissen W, Mailleux J, Nijland PG, Zelcer N, Vanmierlo T, Van Horssen J, Stinissen P, Hellings N, Hendriks JJ. Myelin alters the inflammatory phenotype of macrophages by activating PPARs. Acta Neuropathol Com. 2013;1:43.
Bogie JF, Stinissen P, Hellings N, Hendriks JJ. Myelin-phagocytosing macrophages modulate autoreactive T cell proliferation. J Neuroinflamm. 2011;8:85.
Zhuang W, Zhang L, Zheng Y, Liu B, Ma C, Zhao W, Liu S, Liu F, Gao C. USP3 deubiquitinates and stabilizes the adapter protein ASC to regulate inflammasome activation. Cell Mol Immunol. 2022;19(10):1141–52.
Festa BP, Siddiqi FH, Jimenez-Sanchez M, Rubinsztein DC. Microglial cytokines poison neuronal autophagy via CCR5, a druggable target. Autophagy 2023:1–3.
Houtman J, Freitag K, Gimber N, Schmoranzer J, Heppner FL, Jendrach M. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019;38(4).
Yang L, Gao X, Tian D, Yang W, Xue S, Cao Z, Sun T. Resolvin D2 activates anti-inflammatory microglia via restoring autophagy flux and alleviate neuropathic pain following spinal cord injury in rats. Exp Neurol. 2023;370:114573.
Amin S, Liu B, Gan L. Autophagy prevents microglial senescence. Nat Cell Biol. 2023;25(7):923–5.
Zhao J, Hu B, **ao H, Yang Q, Cao Q, Li X, Zhang Q, Ji A, Song S. Fucoidan reduces lipid accumulation by promoting foam cell autophagy via TFEB. Carbohyd Polym. 2021;268:118247.
Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–67.
Robichaud S, Rasheed A, Pietrangelo A, Doyoung KA, Boucher DM, Emerton C, Vijithakumar V, Gharibeh L, Fairman G, Mak E, et al. Autophagy is differentially regulated in leukocyte and nonleukocyte foam cells during atherosclerosis. Circ Res. 2022;130(6):831–47.
Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15(4):534–44.
Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, Robbins J, Martinez J, Tabas I. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15(4):545–53.
Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, Ali S, Dehestani B, Holloway KB, Micevych PS, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8:15750.
Berglund R, Guerreiro-Cacais AO, Adzemovic MZ, Zeitelhofer M, Lund H, Ewing E, Ruhrmann S, Nutma E, Parsa R, Thessen-Hedreul M et al. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci Immunol. 2020;5(52).
Zhou W, Pal AS, Hsu AY, Gurol T, Zhu X, Wirbisky-Hershberger SE, Freeman JL, Kasinski AL, Deng Q. MicroRNA-223 suppresses the canonical NF-κB pathway in basal keratinocytes to dampen neutrophilic inflammation. Cell Rep. 2018;22(7):1810–23.
He Y, Hwang S, Cai Y, Kim SJ, Xu M, Yang D, Guillot A, Feng D, Seo W, Hou X, et al. MicroRNA-223 ameliorates nonalcoholic steatohepatitis and Cancer by targeting multiple inflammatory and oncogenic genes in hepatocytes. Hepatology. 2019;70(4):1150–67.
Yuan S, Wu Q, Wang Z, Che Y, Zheng S, Chen Y, Zhong X, Shi F. miR-223: an immune regulator in infectious disorders. Front Immunol. 2021;12:781815.
Jiao P, Wang XP, Luoreng ZM, Yang J, Jia L, Ma Y, Wei DW. miR-223: an effective regulator of immune cell differentiation and inflammation. Int J Biol Sci. 2021;17(9):2308–22.
Nguyen MA, Hoang HD, Rasheed A, Duchez AC, Wyatt H, Cottee ML, Graber TE, Susser L, Robichaud S, Berber İ, et al. miR-223 exerts translational control of proatherogenic genes in macrophages. Circ Res. 2022;131(1):42–58.
Wang J, Bai X, Song Q, Fan F, Hu Z, Cheng G, Zhang Y. miR-223 inhibits lipid deposition and inflammation by suppressing toll-like receptor 4 signaling in macrophages. Int J Mol Sci. 2015;16(10):24965–82.
Galloway DA, Blandford SN, Berry T, Williams JB, Stefanelli M, Ploughman M, Moore CS. miR-223 promotes regenerative myeloid cell phenotype and function in the demyelinated central nervous system. GLIA. 2019;67(5):857–69.
Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS, Gerich ME, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214(6):1737–52.
Huppertz C, Jäger B, Wieczorek G, Engelhard P, Oliver SJ, Bauernfeind FG, Littlewood-Evans A, Welte T, Hornung V, Prasse A. The NLRP3 inflammasome pathway is activated in sarcoidosis and involved in granuloma formation. Eur Respir J. 2020;55(3).
Zhou Y, Chen E, Tang Y, Mao J, Shen J, Zheng X, **e S, Zhang S, Wu Y, Liu H, et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019;10(11):843.
Wang H, Chen J, Zhang S, Zheng X, **e S, Mao J, Cai Y, Lu X, Hu L, Shen J, et al. MiR-223 regulates autophagy associated with cisplatin resistance by targeting FBXW7 in human non-small cell lung cancer. Cancer Cell Int. 2020;20:258.
Wu W, Shan Z, Wang R, Chang G, Wang M, Wu R, Li Z, Zhang C, Li W, Wang S. Overexpression of miR-223 inhibits foam cell formation by inducing autophagy in vascular smooth muscle cells. Am J Transl Res. 2019;11(7):4326–36.
Qi X, Man SM, Malireddi RK, Karki R, Lupfer C, Gurung P, Neale G, Guy CS, Lamkanfi M, Kanneganti TD. Cathepsin B modulates lysosomal biogenesis and host defense against Francisella novicida infection. J Exp Med. 2016;213(10):2081–97.
Man SM, Kanneganti TD. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy. 2016;12(12):2504–2505.
Chen X, Yu C, Kang R, Kroemer G, Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28(4):1135–48.
Peng J, Gao C, Chen X, Wang T, Luo C, Zhang M, Chen X, Tao L. Ruxolitinib, a promising therapeutic candidate for traumatic brain injury through maintaining the homeostasis of cathepsin B. Exp Neurol. 2023;363:114347.
Acknowledgements
We acknowledge DIANA-miRPath v4.0 database for providing their platforms and contributors for uploading their meaningful datasets.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81972064 and No.81902217), the Natural Science Foundation of Guangdong Province (2023A1515010565, 2020A1515011415 and 2024A1515010382), Guangzhou Science and Technology Plan Project (202102021244) and the President Foundation of Nanfang Hospital, Southern Medical University (No.2023A024).
Author information
Authors and Affiliations
Ethics declarations
Ethics approval and consent to participate
Animal experiments were carried out by the Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Review Board of Nanfang Hospital, Southern Medical University (NFYY-2017-114).
Consent for publication
All authors have read and agreed with the submission of the manuscript to.
Competing interests
The authors declare no competing interests.
Cell Communication and Signaling. This manuscript has not been presented.
elsewhere entirely.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, H., Ou, Zl., Alaeiilkhchi, N. et al. MiR-223 enhances lipophagy by suppressing CTSB in microglia following lysolecithin-induced demyelination in mice. Lipids Health Dis 23, 194 (2024). https://doi.org/10.1186/s12944-024-02185-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-024-02185-y