Abstract
Background
Considering the essential roles that genetic factors play in azoospermia and oligospermia, this study aims to identify abnormal chromosomes using karyoty** and CNVs and elucidate the associated genes in patients.
Methods
A total of 1157 azoospermia and oligospermia patients were recruited, of whom, 769 and 674 underwent next-generation sequencing (NGS) to identify CNVs and routine G-band karyoty**, respectively.
Results
First, 286 patients were co-analyzed using CNV sequencing (CNV-seq) and karyoty**. Of the 725 and 432 patients with azoospermia and oligospermia, 33.8% and 48.9% had abnormal karyotypes and CNVs, respectively. In particular, 47,XXY accounted for 44.18% and 26.33% of abnormal karyotypes and CNVs, respectively, representing the most frequent genetic aberration in azoospermia and oligospermia patients. Nevertheless, big Y and small Y accounted for 7.46% and 16.67% of abnormal karyotypes, respectively. We also identified high-frequency CNVs-loci, such as Xp22.31 and 2p24.3, in azoospermia and oligospermia patients.
Conclusion
Sex chromosome and autosomal CNV loci, such as Xp22.31 and 2p24.3, as well as the associated genes, such as VCX and NACAP9, could be candidate spermatogenesis genes. The high-frequency abnormal karyotypes, CNV loci, and hot genes represent new targets for future research.
Similar content being viewed by others
Background
Infertility is the inability to initiate a pregnancy after more than one year of regular unprotected sexual intercourse [1]. Approximately 15% of couples are infertile, with male factors accounting for more than 50% of cases. The primary cause of male infertility is abnormal spermatogenesis, such as azoospermia, oligospermia, poor sperm motility, and morphological abnormalities [2,3,4]. Approximately 30% of male infertility cases caused by spermatogenic issues are associated with genetic abnormalities [5]. More specifically, chromosomal abnormalities, chromosomal polymorphisms, sex chromosomal and autosomal microdeletions and microduplications, gene mutations, and mitochondrial genome mutations and deletions that lead to overexpression and deletion negatively impact spermatogenesis, affecting sperm concentration or motility [2, 6,7,8].
Approximately 10% of male infertility patients are categorized as non-obstructive azoospermia (NOA) due to failure in spermatogenesis. Microdeletions and chromosomal abnormalities of the Y chromosome are the key genetic factors in NOA [9], with incidences of 9.7% and 13% in males with azoospermia, respectively [16]. Moreover, CNV-seq has a low associated cost, short run period, high resolution, high accuracy, and requires a small amount of input DNA. Meanwhile, karyoty**, with a maximum resolution of 3 Mb, is the gold standard for identifying chromosomal abnormalities to detect whole and partial chromosomal aneuploidies, unbalanced translocations, polyploidies, and mosaicism [16, 17].
This study analyzed the NGS-based CNV-seq and karyoty** in males with azoospermia and oligospermia to identify novel microdeletion and microduplication targets, as well as genes and abnormal chromosomal karyotypes associated with azoospermia and oligospermia.
Materials and methods
Inclusion criteria
Healthy couples that had not become pregnant after one year of sexual intercourse without contraception, and who showed abnormal semen counts upon semen examination were included in this study.
Exclusion criteria
Adult males with a history of genital tract obstruction or dysfunction (varicocele and obstructive azoospermia) or congenital defects in the structure of the urogenital system were excluded. Adult male patients who had received radiotherapy for reproductive system carcinoma or chemotherapy for malignancies were also excluded.
Semen analysis
All sperm samples were tested using standard procedures in the laboratory department of Tongji Hospital in Wuhan, China. Semen samples were collected through masturbation after 3–5 days of abstinence, and semen characteristics were evaluated within 1 h after ejaculation. Semen analysis was performed in accordance with the World Health Organization (WHO) standard protocol for diagnosis of azoospermia (absence of sperm in semen) and oligospermia (sperm counts < 15 × 106 ml− 1).
Study population
We recruited 1157 male infertility patients with azoospermia or oligospermia from the obstetrics and andrology department of Tongji Hospital, Wuhan, China, between February 2017 and July 2019. All CNV-seq and karyotype analyses of infertile male samples were performed at the perinatal laboratory of Tongji Hospital (Wuhan, China).
Chromosome karyotype analysis of peripheral blood
Approximately 2 ml of peripheral blood was collected in heparin vacutainers for chromosomal karyotype analysis. For each sample, 0.5 ml of peripheral blood was cultured in 5 ml complete medium (RPMI 1640 media + 15% fetal calf serum) for 72 h. The basic cytogenetic method included chromosome harvesting, slide preparation, staining, and banding; subsequently, chromosome number and banding patterns were evaluated. Chromosome preparation involved arresting the cells, harvesting and preparing a single cell suspension, hypotonic treatment, and fixation. The culture process was performed according to standard procedures [18]. G-banded metaphase chromosomes were obtained by treating the cells with trypsin and staining with Giemsa stain. In all cases, at least 30 cells in metaphase were analyzed. In mosaicism-suspected cases, 50 cells were counted. The karyotypes were interpreted according to the International System for Human Cytogenetic Nomenclature (ISHCN) recommendations. An abnormal clone was defined as exhibiting structural aberration, or chromosomal gain in at least two metaphases, and chromosome deletion observed in at least three metaphases.
Sample preparation and copy number variation sequencing (CNV-seq)
For CNV-seq analysis, approximately 1–2 ml of peripheral blood was collected in EDTA vacutainers and sent to the perinatal laboratory of Tongji Hospital, Wuhan, China. DNA was extracted from the peripheral blood using a DNeasy Blood and Tissue Kit (Qiagen, Germany). The DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, United States) (concentration > 50 ng/µl; OD260/280 > 1.8; OD260/230 > 1.5). DNA samples were sent to Bei**g Berry Genomics Co., Ltd., for CNV sequencing using the NextSeq CN500 platform (Illumina Inc.). The annotation and interpretation were carried out based on the guidelines of the American College of Medical Genetics and Genomics. The results were determined by referring to the hg19 version of the human genome and the latest data available from the Database of Genomic Variants (DGV), DECIPHER, Online Mendelian Inheritance in Man (OMIM), University of California Santa Cruz (UCSC), and PubMed.
Genes involved in CNVs
Microdeletion and microduplication of chromosomal fragments detected through CNV-seq may include changes in genetic function and affect gene expression in testicular or other tissues, leading to spermatogenic failure. Hence, the PubMed public data platform of the National Library of Medicine and National Center for Biotechnology Information (NCBI) through Genome Data Viewer was used to search genes of the CNV loci (https://www.ncbi.nlm.nih.gov/genome/guide/human/).
Results
Analysis process
This study recruited 1157 males with infertility, including 725 with azoospermia and 432 with oligospermia. The average ages of patients with azoospermia and oligospermia were 32 ± 0.4 vs. 31 ± 0.6 years (p = 0.57), respectively. The simplified analysis process is shown in Fig. 1. Samples for 769 and 674 patients were evaluated via CNV-seq or chromosome karyoty**, respectively, while those for 286 patients were assessed by both methods. Moreover, 26.92% (77/286) of patients had abnormal CNVs and normal chromosome karyoty**, whereas CNVs were not detected in 7.34% (21/286) of patients that had abnormal chromosome karyoty** (Supplemental Table 1).
Frequency of microduplication and microdeletion CNVs
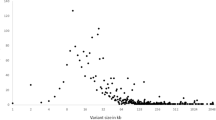
The frequency of CNVs on different chromosomes is shown in Fig. 2. The highest frequency microduplication CNVs occurred in chromosome X, accounting for 36.07% (123/341) of all CNVs. This was followed by chromosomes 2, 11, 7, 4, 12, and Y, accounting for 8.80% (30/341), 5.28% (18/341), 4.69% (16/341), 4.40% (15/341), 4.40% (15/341), and 4.11% (14/341) of CNVs, respectively.
The highest frequency microdeletion CNVs occurred in chromosome Y, which accounted for 26.72% (35/131) of all CNVs. This was followed by chromosomes 14, 4, X, 8, and 10, accounting for 7.63% (10/131), 6.87% (9/131), 6.11% (8/131), 6.11% (8/131), and 5.34% (7/131) of CNVs, respectively.
Abnormal CNVs were detected in 376 patients, of which 52 carried duplication and deletion CNVs, while 324 patients had either duplication or deletion CNVs. No statistical differences were observed between azoospermia and oligospermia patients in the total deletion and duplication CNVs or in the deletion and duplication CNVs of different chromosomes (p > 0.05; Supplementary Table 2).
Analysis of abnormal chromosomal karyoty** in males with azoospermia and oligospermia
Among the males with infertility, 446/674 cases had normal chromosomal karyoty**, while 228 had abnormal karyoty**. Of these 228 cases, the most frequent abnormal karyoty** was 47,XXY, 103/228 (45.18%) in 100 azoospermia cases and 3 oligospermia cases. Small Y (38/228, 20 azoospermia cases, 18 oligospermia cases), big Y (17/228, 5 azoospermia cases, 12 oligospermia cases), abnormal chromosome Y (25/228), and abnormal chromosome X (2/228) were also of interest. The distribution of abnormal chromosomal karyoty** in azoospermia and oligospermia patients is shown in Supplemental Table 3. The 25 cases with karyoty** related to abnormal chromosome Y are shown in Table 1.
High-frequency duplication CNVs
Abnormal CNVs were detected in 376 of the 769 patients. Among these 376, 341 had duplication CNVs, while the others had deletion CNVs (131 patients). Regarding the duplication and deletion CNVs, we focused primarily on chromosomes X, 2, and Y. CNV-seq detected 99 patients with 47,XXY, of which 22 also presented with microduplication and microdeletion of other chromosomes (Supplemental Table 4). Moreover, in the 24 cases of chromosome X microduplication CNVs, 6 cases carried dup(X)(p22.31), 3 dup(X)(p22.33), 2 dup(X)(q21.1), 2 dup(X)(q21.33), 2 dup(X)(q26.3), 2 dup(X)(q27.2), and 2 carried dup(X)(q28). The CNV loci covered genes including VCX, VCX3A, PUDP, SOX3, and HSFX2 (Table 2). The other chromosomal microdeletions and microduplications in chromosome X are presented in Supplemental Table 5. All genes covered by CNV loci are shown in the Supplemental material of Excel 1. Among the 30 cases of chromosome 2 microduplication CNVs, 8 patients carried dup(2)(p24.3), 7 of which had the same NACAP9. Moreover, 3 cases had dup(2)(q32.3) and dup(2)(q37.3), respectively. The CNV loci covered numerous genes, including ANKRD11P1, NRXN1, ITSN, SLC39A10, HES6, AGXT, etc. (Table 3). Other chromosomal microdeletion and microduplications accompanied by chromosome 2 aberrations are shown in Supplemental Table 6. All genes covered by CNV loci are presented in the Supplemental material of Excel 2.
Deletion and microdeletion CNVs on chromosome Y
A total of 131 deletion and microdeletion CNVs were detected in azoospermia and oligospermia patients. However, herein, we focused on 35 patients with deletions in chromosome Y. Of these, 24 cases had AZF region deletion, including all regions of AZF (5/24), AZFa (5/24), AZFb (6/24), AZFc (2/24), and AZFb + AZFc (6/24). Four cases of chromosome mosaicism of 45, X/46, XY were also detected. The clinical manifestation was azoospermia. Three cases were detected with del(Y)(q11.223), of which two were accompanied by other chromosomal microduplications, namely, dup(20)(p12.1) and dup(7)(q36.2), which manifested as azoospermia or oligospermia, respectively. The CNV loci-covered genes included NLGN4Y, TTTY5, DAZ1, and USP9Y. The deletion and microdeletion CNVs and involved genes of chromosome Y are shown in Table 4. All genes covered by CNV loci are shown in the Supplemental material of Excel 3.
Discussion
The WHO estimates that male factors have a crucial role in half of all fertility cases [19]. Male infertility is a complex multifactorial pathological condition that presents as azoospermia and oligospermia. The causes of spermatogenic defects are associated with the environment, lifestyle, nutrition, and genetic factors [20]. In fact, in azoospermia, approximately 25% of cases can be associated with genetic factors; however, the genetic factors of male infertility are highly complex [21].
Somatic cells, germ cells, and endocrine signals are essential for the development and maturation of sperm cells. Both sex chromosomal and autosomal genes are involved in the regulation of spermatogenesis. Cytogenetic karyoty** and NGS-based CNV-seq technologies for chromosomal analysis have been widely used in clinical practice. Chromosomal karyotype analysis is the first choice for genetic testing of infertility in males, especially males with azoospermia and oligospermia. The reason is that the karyotype analysis is simple and convenient, with high accuracy and low cost. In this study, Klinefelter’s syndrome (KS, 47,XXY) was identified as a common chromosomal abnormality in males with infertility (103/674, 15.28%). However, a 45,X/46,XY karyotype was also observed in two males with azoospermia. Chromosomal polymorphisms are primarily considered to have no obvious clinical phenotypes, however, other studies have reported that they are related to infertility and recurrent spontaneous abortion (RSA) [6]. Big Y and small Y are considered Y chromosomal polymorphisms associated with RSA. Hence, we speculate that big Y and small Y could be related to azoospermia and oligospermia. Other chromosome Y abnormalities, such as 47,XYY and 46,X,+mar, and structural aberrations of chromosome Y (inversions, translocations, deletions, and additions), as well as Yqh + and Yqh- were also observed in males with azoospermia and oligospermia. We found a rare case of a male with azoospermia who had a 46,XX karyotype. This testicular disorder of sex development is a rare genetic syndrome, causing male infertility due to the absence of the AZF region in chromosome Yq [22].
Sex chromosomes and autosomal genes participate in spermatogenesis regulation [20]. Meanwhile, 47,XXY (KS) is the main genetic factor of male infertility, accounting for 10–15% of oligospermia and NOA cases [23]. In our study, we detected 99 KS patients through CNV-seq, 77 of which were 47,XXY; 72/77 cases showed azoospermia, and 5 exhibited oligospermia. Furthermore, 22 cases (22/99) were 47,XXY accompanied by other chromosomal microdeletions and microduplications, except for one case of 47,XXY with dup(18)(p11.32) which manifested as oligospermia, while the other 21 cases manifested as azoospermia (Supplemental Table 4). Chromosome X is enriched in genes associated with spermatogenesis. Infertile males reportedly have Xp22.31, Xp22.1, Xp11.23, and Xq28 CNV loci [24]. In this study, 24 patients with chromosome X microduplication CNVs were detected, of which 7 were accompanied by other chromosomal microdeletions and microduplications (Table 2). Moreover, 5 of 6 patients with dup(X)(p22.31) exhibited azoospermia. As such, we speculate that the Xp22.31 loci might be associated with male fertility. The regions involved VCX, VCX3A, GPC3, PUDP, SOX3, and HSFX2, among others. X-linked/Y-linked (VCX/Y) genes encode a group of proteins expressed exclusively in male germ cells among normal tissue. The family members include VCX, VCX2, VCX3A, VCX3B, VCY, and VCY1B. All copies of VCX and VCY are transcribed exclusively in the testis, likely within germ cells [25]. Moreover, the VCX/Y protein family might participate in ribosome assembly regulation during spermatogenesis [26]. VCX may act by mediating the mitochondria-dependent apoptosis pathway or p53-Bax pathway to prompt cell apoptosis and inhibit cell growth, delaying cell progression in G1 to S transition, and leading to spermatogenesis impairment and failure [27]. Some genes have significance in spermatogenesis, while the functions of others are unknown. For example, SOX3 is important for regulating embryonic development and shows homology to the SRY gene. There is enrichment in male-specific genes on the X chromosome [20]. Therefore, the microdeletion and microduplication of CNVs on the X chromosome are responsible for azoospermia and oligospermia. The severity of the clinical phenotype is related to the length of deletion and duplication bases, the location of the region, and whether other chromosomal microdeletions and microduplications are present. In these patients, we detected that the most frequent autosomal CNVs was microduplication on chromosome 2 (30 cases). Moreover, 7/30 cases had dup(2)(p24.3) (chromosome 2: 13,520,000–14,340,000), involving NACAP9, LOC105373438, and LINC00276, among others (Table 3). We considered that the CNV loci of 2p24.3 may be associated with spermatogenesis. Additionally, the 2p24.3 location on chromosome 2 (12,000,001–16,500,000) covered many genes, only a portion of which (including MIR3681HG, MIR3125, LOC100506474, LINC00276, LRATD1, NBAS, DDX1, MYCNUT, MYCNOS, MYCN, and GACAT3) have been previously reported in literature, primarily in association with different cancer types. Meanwhile, only one gene MYCN may be associated with male infertility [28]. However, the roles of NACAP9, LINC00276, and MYCN in spermatogenesis require further investigation. Moreover, given that only 2/4 patients with the same dup(2)(p24.3) loci manifested as azoospermia and oligospermia, spermatogenesis might be associated with factors other than genomics.
The microdeletions in the AZF region on chromosome Y are well-characterized causes of male infertility involving azoospermia and oligospermia [9, 13]. The AZF region contains many critical genes associated with spermatogenesis, germ cell development, and spermatocyte maturation, such as USP9Y, KDM5D, and DAZ [29]. Microdeletion loci on chromosome Y, such as Yp11.2, Yq11.21q11.221, and Yq11.223, may be associated with spermatogenesis. Hence, the associated genes, including NLGN4Y, TTTY5, and SRY, require further functional analyses to clarify their relationship with spermatogenesis. With the rapid development of sequencing technology, genomics has provided new tools to better understand the genetics of male infertility. In particular, deletion and microdeletion of the AZF region of chromosome Y have been found to involve genes related to spermatogenesis. Moreover, microdeletion and microduplication of chromosome X, autosomal genes, involved genes, and abnormal chromosomal karyotypes are associated with spermatogenesis and warrant further research.
The limitations of our study include its small sample size and single research center design. Moreover, we did not perform CFTR analysis nor aCGH and Taqman assays to verify detected CNV loci. Hence, since CNV-seq cannot detect single base mutations and certain microdeletions or microduplications, exome sequencing is needed in future studies.
Conclusion
Various genetic factors contribute to spermatogenic dysfunction. Deletion of the AZF region of the Yq arm primarily manifests as azoospermia and oligospermia. Meanwhile, small Y and big Y may be related to azoospermia and oligospermia. CNV-seq detected that the AZF region of chromosome Y deletion primarily manifested as azoospermia and oligospermia. Various microdeletion and microduplication CNV loci of chromosome Y and the covered genes require further investigation to elucidate their roles in spermatogenesis. Nevertheless, our CNV-seq analysis detected important CNV loci, such as Xp22.31 and 2p24.3, and the associated genes (VCX3A and NACAP9), which might serve as candidate spermatogenesis CNV loci and hot genes.
Data Availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request. Data are located in controlled access data storage at Prenatal Diagnosis Center of Tongji Hospital.
Abbreviations
- CNVs:
-
Copy number variations
- CNV-seq:
-
CNV sequencing
- NGS:
-
Next-generation sequencing
- NOA:
-
Non-obstructive azoospermia
- AZF:
-
Azoospermia factor
- Yq:
-
Long arm of the Y chromosome
- RSA:
-
Recurrent spontaneous abortion
- DGV:
-
Database of Genomic Variants
- OMIM:
-
Online Mendelian Inheritance in Man
References
Waseem AS, Singh V, Makker GC, Trivedi S, Mishra G, Singh K, Rajender S. AZF deletions in indian populations: original study and meta-analyses. J Assist Reprod Genet. 2020;37(2):459–69.
Akınsal EC, Baydilli N, Dündar M, Ekmekçioğlu O. The frequencies of Y chromosome microdeletions in infertile males. Turk J Urol. 2018;44(5):389–92.
Iijima M, Shigehara K, Igarashi H, Kyono K, Suzuki Y, Tsuji Y, Kobori Y, Kobayashi H. Mizokami A.Y chromosome microdeletion screening using a new molecular diagnostic method in 1030 japanese males with infertility. Asian J Androl. 2020;22(4):368–71.
Hwang K, Yatsenko AN, Jorgez CJ, Mukherjee S, Nalam RL, Matzuk MM, Lamb DJ. Mendelian genetics of male infertility. Ann N Y Acad Sci. 2010;1214:E1–E17.
Liu CL, Zhao XY, Mu CL, Li H, Ma J, Jiao HY, Huo ZH. The Association of partial azoospermia factor C deletions and male infertility in Northwestern China. Hum Hered. 2019;84(3):144–50.
Yan JH, Fan LL, Zhao YR, You L, Wang LC, Zhao H, Li Y, Chen ZJ. DYZ1 copy number variation, Y chromosome polymorphism and early recurrent spontaneous abortion/early embryo growth arrest. Eur J Obstet Gynecol Reprod Biol. 2011;159(2):371–4.
Faja F, Carlini T, Coltrinari G, Finocchi F, Nespoli M, Pallotti F, Lenzi A, Lombardo F, Paoli D. Human sperm motility: a molecular study of mitochondrial DNA, mitochondrial transcription factor A gene and DNA fragmentation. Mol Biol Rep. 2019;46(4):4113–21.
Schoot VV, Viellevoije SJ, Tammer F, Brunner HG, Arens Y, Yntema HG, Oerlemans AJ. The impact of unsolicited findings in clinical exome sequencing, a qualitative interview study. Eur J Hum Genet. 2021;29(6):930–9.
Matsumura T, Endo T, Isotani A, Ogawa M, Ikawa M. An azoospermic factor gene, Ddx3y and its paralog, Ddx3x are dispensable in germ cells for male fertility. J Reprod Dev. 2019;65(2):121–8.
Dong Y, Pan Y, Wang R, Zhang Z, ** Q, Liu RZ. Copy number variations in spermatogenic failure patients with chromosomal abnormalities and unexplained azoospermia. Genet Mol Res. 2015;14(4):16041–9.
Sha YW, Mei LB, Ji ZY, Ding L, Ge YS, Wu Q, Kong H, Su ZY, Li P. Two cases of complex balanced autosomal translocations associated with severe oligozoospermia. Gene. 2018;663:126–30.
Khiavi MA, Jalili A, Safary A, Gharedaghchi Z, Mirinezhad SK, Mehdizadeh A, Rahmani SA. Karyotypic abnormalities and molecular analysis of Y chromosome microdeletion in iranian Azeri turkish population infertile men. Syst Biol Reprod Med. 2020;66(2):140–6.
Nailwal M, Chauhan JB. Azoospermia factor C subregion of the Y chromosome. J Hum Reprod Sci. 2017;10(4):256–60.
Huang W, Wang J, Pang M, Zhao Q, Kong L, Mao Y, Li W, Liang Β. Copy number variations in female infertility in China. Balkan J Med Genet. 2019;22(1):5–10.
Zhao XX, Fu L. Efficacy of copy-number variation sequencing technology in prenatal diagnosis. J Perinat Med. 2019;47(6):651–5.
Ma N, ** and SNP-array analyses for effective prenatal diagnosis of chromosomal mosaicism. BMC Med Genomics. 2021;14(1):56.
Liu S, Song L, Cram DS, ** vs copy number variation sequencing for detection of chromosomal abnormalities associated with spontaneous miscarriage. Ultrasound Obstet Gynecol. 2015;46(4):472–7.
Bates SE. Classical cytogenetics: Karyoty** techniques. Methods Mol Biol. 2011;767:177–90.
Fainberg J, Kashanian JA. Recent advances in understanding and managing male infertility. F1000Res. 2019; 8:F1000 Faculty Rev-670.
Singh K, Jaiswal D. Human male infertility: a complex multifactorial phenotype. Reprod Sci. 2011;18(5):418–25.
Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15(6):369–84.
Li TF, Wu QY, Zhang C, Li WW, Zhou Q, Jiang WJ, Cui YX, **a XY, Shi YC. 46,XX testicular disorder of sexual development with SRY-negative caused by some unidentified mechanisms: a case report and review of the literature. BMC Urol. 2014;14:104.
Sciarra F, Pelloni M, Faja F, Pallotti F, Martino G, Radicioni AF, Lenzi A, Lombardo F, Paoli D. Incidence of Y chromosome microdeletions in patients with Klinefelter syndrome. J Endocrinol Invest. 2019;42(7):833–42.
Krausz C, Giachini C, Giacco DL, Daguin F, Chianese C, Ars E, Ruiz-Castane E, Forti G, Rossi E. High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PLoS ONE. 2012;7(10):e44887.
Jakobsen MK, Traynor S, Stæhr M, Duijf PG, Nielsen AY, Terp MG, Pedersen CB, Guldberg P, Ditzel HJ, Gjerstorff MF. The Cancer/Testis Antigen Gene VCX2 is rarely expressed in Malignancies but can be epigenetically activated using DNA methyltransferase and histone deacetylase inhibitors. Front Oncol. 2021;10:584024.
Zou SW, Zhang JC, Zhang XD, Miao SY, Zong SD, Sheng Q, Wang LF. Expression and localization of VCX/Y proteins and their possible involvement in regulation of ribosome assembly during spermatogenesis. Cell Res. 2003;13(3):171–7.
Ji J, Qin Y, Wang R, Huang Z, Zhang Y, Zhou R, Song L, Ling X, Hu Z, Miao D, Shen H, **a Y, Wang X, Lu C. Copy number gain of VCX, X-linked multi-copy gene, leads to cell proliferation and apoptosis during spermatogenesis. Oncotarget. 2016;7(48):78532–40.
Xu W, Hu H, Wang Z, Chen X, Yang F, Zhu Z, Fang P, Dai J, Wang L, Shi H, Li Z, Qiao Z. Proteomic characteristics of spermatozoa in normozoospermic patients with infertility. J Proteom. 2012;75(17):5426–36.
Zhu Y, Hu LS, Cao DL, Ou XH, Jiang MX. Chromosomal microarray analysis of infertile men with azoospermia factor microdeletions. Gene. 2020;735:144389.
Acknowledgements
We would like to express our thanks to all people who participated in this study. We also thank Editage for scientific editing of this manuscript.
Funding
This study was funded by National Natural Science Foundation of China and Bei**g Kanghua Western and Chinese Medicine Development Foundation (Grant No. 81902299 & 2201102997).
Author information
Authors and Affiliations
Contributions
XX, SW, and LF conceived and designed the study, analyzed the data, and drafted and revised the paper. XP, YJ, NW, JZ, SL, and HH prepared and analyzed the data. YZ, CZ, LZ helped to revise the tables. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in this study were in accordance with the ethical guidelines of the ethics committee of Tongji Medical College, Huazhong University of Science and Technology. All study participants provided written informed consent. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-023-01652-2