Abstract
DNA topoisomerases regulate conformational changes in DNA topology during normal cell growth, such as replication, transcription, recombination, and repair, and may be targeted for anticancer drugs. A DNA topology assay was used to investigate DNA-damaging/protective activities of extracts from Habanero Red (HR), Habanero Maya Red (HMR), Trinidad Moruga Scorpion (TMS), Jalapeno (J), Serrano pepper (SP), Habanero Red Savina (HRS), Bhut Jolokia (BJ), and Jamaica Rosso (JR) peppers, demonstrating their inhibitory effect on the relaxation of pBR by Topo I. DNA topoisomerase II (Topo II) is proven therapeutic target of anticancer drugs. Complete inhibition of Topo II was observed for samples TMS, HR, and HMR. Extracts J and SP had the lowest capsaicin and dihydrocapsaicin content compared to other peppers. HR, HMR, TMS, J, S, HRS, BJ, JR extracts showed the anticancer effect, examined by MTS and xCell assay on the in vitro culture of human colon carcinoma cell line HCT116.
Similar content being viewed by others
Introduction
Cancer disease is a serious health and social problem. Despite therapeutic advances, cancer is the second leading cause of morbidity and mortality worldwide (https://www.who.int/health-topics/cancer#tab=tab_1). Although treating cancer with chemotherapy and radiotherapy is effective, it is associated with serious side effects, such as drug resistance or non-selectivity [1]. These problems illustrate the need to develop new, more effective anticancer therapies and safer agents [2]. Natural products or their direct derivatives play an important role in the discovery of new drugs for the treatment of cancer [3]. The plant compounds have different inhibitory effects on cancer onset, development, progression, and metastasis [4, 5]. Plants of the genus Capsicum, belonging to the family Solanaceae, are an important source of biologically active substances [6]. We currently recognize 25 wild species and five domesticated species in the genus Capsicum [7], Capsicum annuum, Capsicum frutescens, Capsicum chinense, Capsicum baccatum, and Capsicum pubescens [8]. In addition to their use in gastronomy, peppers are also excellent producers of secondary metabolites, which have various pharmacological properties and contain cytotoxic compounds [9].
A characteristic feature of wide varieties of peppers is their intense pungency caused by a group of bioactive phytochemicals, capsaicinoids, classified as alkaloids. They are vanilylamides derived from branched-chain C8-C11 (E) -monocline fatty acids and branched-chain or straight-chain saturated fatty acids [10]. One of the capsaicinoids, capsaicin (48.6%) (CAP) is the most abundant compound in chili peppers, followed by 6,7-dihydrocapsaicin (36%) (DHK), nordihydrocapsaicin (7.4%), homodihydrocapsaicin (2%), and homocapsaicin (2%). CAP (trans-8-methyl-N-vanillyl-6-non enamide) is a crystalline, lipophilic, colorless, and odorless alkaloid soluble in fats, alcohols, and oils [11, 12]. Many studies have shown that capsaicinoids have a wide range of biological and physiological effects. Capsaicinoid biosynthesis and accumulation is a genetically determined trait in chili pepper fruits as different cultivars or genotypes, where gene expression has identified candidate genes possibly involved in capsaicinoid biosynthesis [10, 13]. CAP has analgesic [14, 15] and anti-inflammatory effects [16], decreases the prevalence of obesity [17] and metabolic syndrome, improves gastrointestinal [18] and cardiovascular symptoms [19], and is characterized by antitumor activity [20, 21]. The nutritional, and anti-obesity properties of different chili peppers was presented by Azlan et al. [22]. Due to ability of CAP to mediate cell cycle arrest and induce cell apoptosis in in vitro experiments, it reduced the growth of human leukemia cells [23], skin tumor cells [24], prostate [25, 26], bladder [27], stomach [28, 29], colon [30], nasopharynx [31], liver [32], lung [33], and breast cancer [34]. Capsaicin can modify the function of many genes associated with the lifespan of cancer cells, initiating apoptosis, arresting cell growth, and suppressing angiogenesis and metastasis [35, 36]. By inducing apoptosis in cancer cell lines, healthy cells remain intact [37]. Several studies have shown that new combination therapies with various phytochemicals and chemopreventive drugs can induce increased antitumor activity through an additive or synergistic effect [38]. Capsaicinoids potentiate the chemotherapeutic effect and relieve pain in cancer patients. CAP acts synergistically with other anticancer agents; thus, it can be used with other chemotherapeutic agents in cancer treatment [39,40,41]. Colorectal cancer is one of the most commonly diagnosed diseases in the world [42]. The incidence of this disease is closely related to the composition of the diet and the amount of vegetables consumed. Currently, preclinical studies testing the anticancer effects of CAP on colon cancer are lacking [43,44,45].
DNA topoisomerases regulate conformational changes in DNA topology during normal cell growth, such as DNA replication, transcription, recombination, and DNA repair [46]. They are also targets for several anticancer drugs [47, 48]. Topoisomerase inhibitors interfere with human topoisomerases, or they can act as inhibitors without tumor cell toxicity [49].
The aim of our work was to investigate the DNA-damaging/protective activities of the studied chili extracts. The DNA topology was studied with electrophoretic detection of topological changes induced in plasmid DNA. We hypothesized that an extract of CAP, DHK, and other varieties of chili peppers would influence both Topo I and II and exploit the ability to act on two distinct enzymatic targets, thereby maximizing the potential therapeutic effects. The biological activity of extracts was assessed using an MTT assay of the human colon cancer cell line HCT-116, potentially usable in cancer therapy and drug screening.
Materials and methods
Sample processing
All analyzed types of chili peppers: Habanero Red (HR), Habanero Maya Red (HMR), Trinidad Moruga Scorpion (TMS), Jalapeno (J), Serrano pepper (SP), Habanero Red Savina (HRS), Bhut Jolokia (BJ), Jamaica Rosso (JR) were grown and harvested at the Department of Food Hygiene, Technology and Safety of the University of Veterinary Medicine and Pharmacy in Košice. The cultivated peppers were dried in a laboratory oven with ventilation at 40 ± 5 °C. Before drying, the chili peppers were cut in half or quarters (depending on the size) in order to speed up the drying and to avoid undesired changes. The peppers were dried together with the placenta and seeds. After drying, they were stored in a closed glass container in a dry, dark place until analysis.
Extraction of capsaicinoids
Completely dried fruits of various varieties of peppers were ground completely on an electric stainless steel mixer. From each pepper sample 0.2 g of ground pepper was weighed into 10 mL volumetric flasks and 2 ml of 96 (v/v) ethanol were added. The mixture was mixed on a Vortex homogenizer and placed in an ultrasonic bath for 5 minutes. After homogenization, the samples were macerated for 24 hrs in a dry and dark place under laboratory conditions. After the indicated time, the individual samples were filtered through filter paper into 10 mL volumetric flasks, washed with absolute ethanol and made up to 10 mL with the same solvent. The extracts were stored in sealed flasks at 5 °C in a refrigerator. Samples were filtered through a membrane syringe filter (Q-Max® RR Siringe Filters, Frisenette, 25 mm, 0.22 μm PVDF) prior to HPLC analysis. If CAP or DHK concentrations were outside the calibration range, the samples were diluted with absolute ethanol.
HPLC analysis
CAP and DHK standards were purchased from Sigma-Aldrich (USA), absolute ethanol from Emparta (Germany) and HPLC grade acetonitrile from Fisher (UK). The concentration of CAP and DHK in the extracts was determined using a Dionex UltiMate 3000 RS with a diode array detector (DAD) and a programmable Chromeleon Chromatography Data System, version 7.2 (Thermo Fisher Scientific, Germany). HPLC analysis was performed using a Polaris 5 column (C18-A 250 × 4.6 mm, 5 m, under isocratic conditions, at 40 °C and flow rate 1 mL.min−1. The sample was dosed using an autosampler and its volume was 10 L. The mixture of acetonitrile and water (70:30, v/v) was used as the mobile phase. CAP and DHK were measured with UV detector (DAD) at 282 nm. The quantification and HPLC method validation was based on the calibration curve fitting by linear regression analysis. Linear correlation between the peak area and the applied concentration was found in the concentration range 5–500 μg.mL−1, as confirmed by the correlation coefficient (0.99902 for CAP and 0.99932 for DHK). The x-axis in the graphical dependence represented the concentration of CAP or DHK and the y-axis was the peak area in the chromatographic record. The mean values for the regression equation were y = 0.027.x + 0.2049 for CAP and y = 0.0067x + 0.0057 for DHK.
Nuclease activity
A nuclease activity study was performed prior to the experiments, which confirmed that none of the samples were able to cleave plasmid DNA and that the ethanol content did not affect the plasmid. Nuclease activity of selected molecules were studied using isolated plasmid pUC19 (isolated by the alkaline lysis method in our laboratory). Mixture of pUC19 in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0, 2 μl) (Sigma-Aldrich), 10 mM Tris-HCl buffer (pH 7.4, 25 μL) (Sigma-Aldrich) and studied compounds (3 μL) in final concentration 1/10 of stock solution were incubated at 37 °C for 18 hrs. After incubation solution of bromophenol blue and xylene violet (3 μL) and samples were subjected on 1.0% v/v agarose (Sigma-Aldrich) gel. Electrophoresis ran 4 h at 35 V in 1xTAE (40 mM Tris, 20 mM acetic acid glacial (Centralchem), 1 mM EDTA) (Sigma-Aldrich) then it was stained with ethidium bromide for 15 min and destained in deionized water for 7 min. Electrophoretic record was photographed with electrophoretic system SYNGEN and processed with GeneSnap program.
Decatenation assay for topoisomerase II
Topoisomerase II (Topo II) decatenation assay was carried out according to the Inspiralis protocol using kinetoplast DNA (kDNA, 200 ng) in TE buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA) (Sigma-Aldrich) and appropriate amount of diluted human topoisomerase IIa (hTop IIa, 1.5 U) enzyme in dilution buffer (50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 50% v/v glycerol, 50 μg/mL albumin (Inspiralis). Assay was conducted in final concentration 1/10 of stock solutions (3 μL from stock solutions, in the case of CAP we used two stock solutions – 0.5 mg/mL and 1.0 mg/mL). Samples were prepared using a mixture of appropriate assay buffer (50 mM Tris-HCl (pH 7.5), 125 mM NaCl, 10 mM MgCl2, 5 mM DTT, 100 μg/mL albumin, supplied as 10 × (Inspiralis)), 30 mM ATP (final concentration 1 mM) and deionized water in final volume 30 μL. Samples were incubated 30 min at 37 °C and after that reaction was stopped with STEB (30 μL, 40% v/v sucrose (Centralchem), 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5 g/dm3 bromophenol blue (Sigma-Aldrich)) and purified with chloroform:isoamylalkohol (Centralchem) (30 μL, 24:1) solution and subjected to the 1% v/v agarose gel in 1× TAE (40 mM Tris, 20 mM acetic acid glacial (Centralchem), 1 mM EDTA (Sigma-Aldrich)). Dilution of ethanol (% v/v) in samples for Topo I a Topo II was 3 μl ethanol/ 30 μl sample. Electrophoresis ran 4 hrs at 35 V and then agarose gel was stained with ethidium bromide solution and destained in water. Electrophoretic record was documented using UV light.
Relaxation assay for topoisomerase I
Impact of molecules on relaxation ability of topoisomerase I was studied on human topoisomerase I (hTopo I, Inspiralis) on plasmid pBR322 (Inspiralis). Mixture of plasmid (0.5 μg) in TE (10 mM Tris-HCl (pH 7.5), 1 mM EDTA), diluted hTopoI (0.5 U) in dilution buffer (10 mM Tris-HCl (pH 7.5), 1 mM DTT, 1 mM EDTA, 50% v/v glycerol, 50 μg/mL albumin (Inspiralis)) and studied compounds in final concentrations of 1/10 of stock solutions (3 μL from stock solution, in case of CAP were used two stock solutions – 0.5 mg/mL and 1.0 mg/mL) were incubated at 37 °C for 30 minutes in 1 × concentrated assay buffer (20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.25 mM EDTA, 5% glycerol, 50 μg/mL albumin, supplied as 10× stock (Inspiralis)) and deionized water in the final volume of 30 uL. After incubation reaction was stopped with STEB (30 μL, 40% v/v sucrose (Centralchem), 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5 g/dm3 bromophenol blue (Sigma-Aldrich)) and samples were purified with chloroform:isoamyl alcohol (Centralchem) (30 μL, 24:1) and upper layer of samples was subsequently subjected on 1% v/v agarose gel. Electrophoresis ran for 15 min at 20 V to allow subjected samples to penetrate the gel and then continued for 4 hrs at 35 V in 1 × TAE buffer (40 mM Tris, 20 mM acetic acid glacial (Centralchem), 1 mM EDTA (Sigma-Aldrich)). Then agarose gel was stained with ethidium bromide solution (15 min) and destained with deionized water (10 min). Electrophoretic record was visualized by UV light, photographed by SYNGEN system and processed in GeneSnap program.
Cell line
Human colon carcinoma cell line HCT116 (ATCC® CCL-247™) was cultured in RPMI medium supplemented by antibiotics (100 U/ml penicillin + 100 μg/mL penicilin-streptomycin) and 10% of FBS (fetal bovine serum) in the presence of 5% CO2 in a humidified atmosphere at 37 °C. If 5 × 106 cells were plated onto a 75 cm2 flask, the culture reaches 70-90% confluency in 2-3 days and was ready to split or harvest for experiments. To determine the linear range of each assay, six cell densities ranging from 50 to 10 000 cells/well were plated into sterile 96-well plates and incubated for 24, 48 or 72 hrs.
MTS assay
MTS assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl) -2H-tetrazolium) as indicators of metabolically active mitochondria overestimated the number of viable cells. MTS was used for determining the number of viable cells in proliferation, cytotoxicity, or chemosensitivity. HCT116 cells were seeded at 5000 cells per well into 96-well microplates, after 24 hrs incubation treated with extracts of individual samples (extracts of peppers), CAP or DHK. After 24 hrs incubation of HCT116 cells, medium was removed and replaced with RPMI containing 10% fetal bovine serum (FBS) and extracts of dry extracts in ethanol peppers: HR, HMR, TMS, J, S, HRS, BJ, JR in dilution 100×, 500×,1000× and incubated at 37 °C and 5% CO2 for 24 hrs. Control group (cells HCT116) was not affected by extracts, CAP1 - CAP6 concentrations of CAP at 10 μM, 25 μM, 50 μM, 100 μM, 150 μM, 200 μM. DHK1 - DHK6 concentrations of DHK at 10 μM, 25 μM, 50 μM, 100 μM, 150 μM, 200 μM. The MTS assay was performed at 48 hrs and 72 hrs. After the 24 hrs exposure to the cells, 25 μL of CellTiter 96, AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Madison, WI, USA) was added to the cell culture medium, and incubated for 3 hrs at RT. The absorbance of wells at 490 nm was measured using a microplate reader. Results were expressed as means (±sd) of quadruplicate wells obtained by subtraction from cell-free equivalents, to eliminate A490 produced by the media alone. Effects of pepper extracts on HCT116 cells were analysed by MTS assay expressed as a fold of control [%] of absorbance generated in cell-mediated MTS assays to the control group.
Agilent × CELLigence real-time cell analysis
HCT116 cells (5 × 103 cells/well) were seeded in 96-well plates (RTCA E-Plates 96) on xCELLigence RTCA systems (Agilent). The cells were treated with chilli extracts 24 hrs after seeding. HCT116cells were cultured in the absence or presence of tested drugs at concentrations ranging from 100 μM to 100 nM. The cell adhesion and spread without the manipulation of the cells were continuously monitored in 60 min intervals over the course of a 120 hrs observation period using the ×CELLigence RTCA system.
Statistical analysis
Experiments under all conditions were performed in at least three independent measurements. Mean value and standard deviation were calculated using descriptive statistics. The data were analyzed by using the RTCA software Pro 1.2.1 (ACEA Bioscience). Statistical analysis was carried out by a non-parametric method, one-way ANOVA using SigmaPlot (Ver. 12.0). Differences were considered significant *p < 0.05; **p < 0.01; ***p < 0.001.
Results and discussion
Capsaicin and dihydrocapsaicin content in chili peppers
The exact content of CAP and DHK in presented different types of chili peppers has not yet been described in the literature; therefore, our results provide novel data. The findings showed the highest content of CAP and DHK in species TMS and BJ, while J and SP peppers had the lowest concentrations of CAP and DHK (Table 1). From the preparation of the experiment, the same conditions, including the same composition of soil and water, were maintained for the growth of pepper plants in the defined conditions. Every single aspect, such as differences in soil composition and chemical composition of water when cultivating plants in different ecological conditions of different areas of the world, could affect the content of individual components in plants. Environmental influences can also be considered the epigenetic factors that play a role in the expression of genes responsible for producing individual components, such as CAP, flavonoids, and phenolics. CAP, as a cancer preventive agent, shows wide applications against various types of cancer [50]. Studies have already determined its antiproliferative activity against HT-29 colon cancer cells and HepG2 liver cancer cells and high antioxidant activity and found high concentrations of CAP, flavonoids, phenolics, and total soluble solids content in nine different peppers belonging to Capsicum annuum (No. 1072 and cultivars CM334, A44750157, PM 217, Sunset, Grif 9285, PM 687) and C. chinense (CA4 and No.1745). Line 1745 of C. chinense showed potential as a nutraceutical compound for the prevention and treatment of colon and liver cancers. The different levels of anticancerous phytochemicals, such as CAP and flavonoids, were also detected in cultivars [51]. The data indicate chili peppers with significantly higher CAP and DHK levels.
Chili extracts and nuclease and topoisomerase activity
Nuclease activity
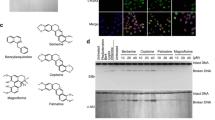
A nuclease activity study confirmed that none of the samples could cleave plasmid DNA and that the ethanol content did not affect the plasmid (Fig. 1).
Nuclease activity. Nuclease activity of selected molecules were studied on isolated plasmid pUC19 and chili extracts in final concentration 1/10 of stock solution incubated at 37 °C for 18 hrs. Compounds of extract: HR – Habanero Red; HMR – Habanero Maya Red; TMS – Trinidad Moruga Scorpion; J – Jalapeno; SP – Serrano pepper; HRS – Habanero Red Savina; BJ – Bhut Jolokia; JR – Jamaica Rosso (final concentration 1/10 of stock solution); CAP – capsaicin (final concentration 0,05 mg/mL); DHK – dihydrocapsaicin (final concentration 0,05 mg/mL); K1 – pUC19; K2 – pUC19 + EtOH
Decatenation assay for topoisomerase II
Human DNA Topo II is a nuclear enzyme that catalyzes the introduction of topological changes to the DNA molecule. hTopo II is effective in treating a wide spectrum of cancers. In the last decade, many scientists have designed, synthesized, and evaluated various bioactive molecules that target Topo II. In our experiment, we used a decatenation assay to measure Topo II’s catalytic activity to decatenate kinetoplast-catenated DNA (kDNA) in a cell-free system.
At the beginning of the experiment, we first looked at whether ethanol (solvent) affects topo IIa and we found that it does not affect the activity of topo II topoisomerase (data not shown). However, a small non-significant effect of ethanol on the activity of topoisomerase IIa cannot be excluded, since the influence of ethanol on topoisomerase activity was demonstrated for topoisomerase I (Figs. 3 and 4). Figure 2 shows the inhibition of Topo II in all samples (Fig. 2), but complete inhibition was observed only for TMS, HR, and HMR. Samples J and SP had the lowest content of CAP and DHK substances, although they had approximately the same inhibition effect as in cases of CAP1 and CAP2 and DHK, suggesting that other substances present in the chili peppers might participate at inhibition activity of Topo II. Nevertheless, the presence of CAP and DHK is important in inhibiting the activity of this enzyme. As we observed a higher amount of catenated DNA in the wells of some samples, we assumed that ethanol could still affect the activity of Topo II.
Decatenation assay for topoisomerase II. Topoisomerase II decatenation assay was carried out using kinetoplast DNA (kDNA, 200 ng) in human topoisomerase IIa (hTop IIa, 1.5 U) enzyme, in 1/10 of stock solutions (3 μL from extracts, capsaicin 0.5 mg/mL and 1.0 mg/mL). Extracts: HR – Habanero Red; HMR – Habanero Maya Red; TMS – Trinidad Moruga Scorpion; J – Jalapeno; SP – Serrano pepper; HRS – Habanero Red Savina; BJ – Bhut Jolokia; JR – Jamaica Rosso (final concentration 1/10 of stock solution); CAP 1 – capsaicin (final concentration 0.05 mg/mL); CAP 2 – capsaicin (final concentration 0.1 mg/mL); DHK – dihydrocapsaicin (final concentration 0.05 mg/mL); K1 – kDNA; K2 – kDNA + Topo II (1.5 U) + EtOH
Relaxation assay for topoisomerase I
The primary reaction of Topo I is the relaxation of supercoiled DNA which has a different electrophoretic mobility than a completely relaxed DNA. The effect of molecules on the relaxation ability of topoisomerase I was studied with human Topo I on plasmid pBR322. The TMS sample completely inhibited Topo I, while no activity was observed for CAP and DHK in the given conditions. For that reason, in the next step, we changed the hTopo I concentration from 0.5 U to 1.0 U (Figs. 3 and 4). We can observe that samples TMS, J, and BJ had the highest ability to inhibit Topo I (despite the weak effect of ethanol on Topo activity).
Relaxation assay for topoisomerase I. HR – Habanero Red; HMR – Habanero Maya Red; TMS – Trinidad Moruga Scorpion; J – Jalapeno; SP – Serrano pepper; HRS – Habanero Red Savina; BJ – Bhut Jolokia; JR – Jamaica Rosso (final concentration 1/10 of stock solution); CAP 1 – capsaicin (final concentration 1/10 of stock solution 0.05 mg/mL); CAP 2 – capsaicin (final concentration 0.1 mg/mL); DHK – dihydrocapsaicin (final concentration 0,05 mg/mL); K1 – pBR322; K2 – pBR322 + Topo I (1.0 U); K3 – pBR322 + Topo I + EtOH
In contrast, SP, HR, and JR had no or little inhibitory activity, as they did not differ from K3 (ethanol probably played a role as a Topo inhibitor). The activity was also observed with CAP and DHK, with stronger inhibition of DHK. Based on the results, the TMS sample appeared to be the best candidate for further investigation, but it should also be noted that this sample contained much higher concentrations of CAP and DHK (approx. 800 μM and 900 μM) than other substances required for inhibition (as an example, acridine derivatives require less than 100 μM, ethidium bromide approximately 10 μM for Topo I, for acridines below 100 μM, and amsacrine below 250 μM for Topo II).
Based on the previous studies [49, 52, 53] class I (Topo poisons) and class II (catalytic inhibitors) DNA topoisomerase inhibitors were described, although not all showed toxicity to tumor cells. Many DNA Topo (class I) inhibitors are already commonly used in antitumor therapy: doxorubicin [54], cisplatin [55], and genistein [56]. The DNA Topo (class II) inhibitors, a plant alkaloid Camptothecin [57], its derivatives topotecan (Hycamtin) and irinotecan (CPT-11, Campostar), and another coumarin Topo inhibitor [58], are currently used in the clinic. Unfortunately, the clinical use of some of these anticancer drugs is limited due to their dose-limiting toxicity and chemical instability [48]. The non-camptothecin hTopo I inhibitors, indolocarbazoles (NB-506 and his derivative Edotecarin (J-107088), indenoisoquinolines (NSC 314622), indotecan (LMP400) and indimitecan (LMP776), and dibenzonaphthyridinones, have been investigated and under clinical development. These drugs have limitations due to their instability, severe side effects, and drug resistance caused by P-glycoprotein [59]. The catalytic activities of topoisomerases are modulated through their interactions with various proteins. The discovery of new anticancer drugs is important for many cancer patients resistant to specific drugs. DNA Topo remains an important therapeutic target of anticancer agents and antibacterial drugs [48]. We analyzed the nuclease activity, decatenation assay for Topo II, and relaxation assay for Topo I and found that the inhibition of DNA Topo I and IIa supported the use of pepper mix as a potential anticancer drug.
Cytotoxic effect of chili extracts on HCT116 cells
The MTS method for the sensitive quantification of viable cells was used to assess the effect of chili extracts on HCT116 cell metabolic activity and/or viability. When comparing the cytotoxic effect of the pepper extracts with the effect of pure CAP and DHK on HCT116 cells, pure CAP and DHK had a comparable effect on cell viability (Fig. 5). TMS extract and CAP5 (150 μM) supported temporarily the viability of HCT116 cells in the observed time up to 48 hrs after administration of the substances to the cells. On the contrary, both TMS and CAP5 extracts reduced the viability of these cells at 72 hrs. In this regards, HR, HMR, J, S, HRS, BJ, JR extracts decreased tumor cell viability after treatment administration, demonstrated at 48 and 72 hrs in vitro.
MTS Analysis. MTS analysis of cell viability at 48 and 72 hrs upon treatment with chili extracts (HR, HMR, TMS, J, S, HRS, BJ, JR) and capsaicin (CAP), dihydrocapsaicin (DHK) in different concentrations cultivated with HCT116 cells. The extracts of peppers HR, HMR, TMS, J, SP, HRS, BJ, JR. Control group (cells HCT116) was not affected by extracts, Cap1 - Cap6 concentrations of CAP at 10 μM, 25 μM, 50 μM, 100 μM, 150 μM, 200 μM. DHK1 - DHK6 concentrations of DHK at 10 μM, 25 μM, 50 μM, 100 μM, 150 μM, 200 μM. MTS analysis expressed as a fold of control [%] of absorbance generated in cell-mediated MTS assays to control group. Absorbance values are obtained from three independent experiments (n = 3 for each group) and values are expressed in mean ± SE. The groups treated with extracts alone were compared with control: * p < 0.05, **p < 0.01, ***p < 0.001. * vs Ctrl, ▲ vs HR and each extract mutually at both analyzed times (48 hrs, 72 hrs). Statistically insignificant [ns]: CAP1 vs DHK1, HR vs DHK3 at 48 hrs; CAP1 vs DHK1, BJ vs CAP3, HR vs DHK3, HRS vs JR at 72 hrs
The effects of extracts of TMS pepper (the content of CAP 122,30 ng/g) were comparable to that of CAP5. Other extract components likely contribute to the resulting biological effect on tumor cells. During the next 24 hrs of incubation, 72 hrs after adding the extracts to the cells, the effect of the extracts of all types of peppers, CAP (CAP 1 – 5), and DHK (DHK 1 – 5) significantly decreased the viability of HCT116 cells. Finally, the MTS assay demonstrated that chili extracts affect the cell viability of cancer cells at the concentrations used. Further analyses need to confirm these results by testing the antitumor effect of the individual components of the pepper extracts, mainly flavonoids and the contents of others substances.
Antiproliferative effect of chili extracts on HCT116 cells
Cell culture assay (xCELLigence systems) showed the effect of all tested chili extracts on the suppression of the growth and proliferation of HCT116 cells at the end of the monitoring period (Fig. 6).
xCell proliferation. HCT116 cells (Ctrl) seeded in plates (RTCA E-Plates 96) were treated with 100 μM chilli extracts 24 hrs after seeding. The cell adhesion and spread were monitored over 90 hrs using the ×CELLigence RTCA system (Agilent). HRS and JR extracts (CAP content (362.57 and 343.40 μg/ml) increased cell proliferation during 30-50 hrs of culture
Despite the inconsistent effect of chili extracts in the first hours after their administration finally, the cell growth and/or proliferation decreased at a low level after treatment of each chili variety. HRS and JR extracts with a moderately high CAP content (362.57 and 343.40 μg/mL) increased cell proliferation during the first 30-50 hrs of culture, probably due to the influence of other components of the extract than the CAP content alone. Conversely, TMS and BJ extracts with higher CAP and DHK contents significantly reduced the proliferation of cancer cells several hours after the treatment. The results revealed TMS, J, and BJ pepper extracts have the highest ability to inhibit Topo I and cell proliferation, implying that they have the potential for medicinal treatment.
Recent studies have shown that pure CAP has antiproliferative and pro-apoptotic effects on different cancer cell lines and found an association of CAP at high doses with mutagenicity and carcinogenicity [60,61,62]. The research on pepper seed extract reported that it supresses the proliferation of human breast cancer cells MDA-MB-231 and MCF-7 cells [63]. CAP administration reduced cell proliferation and modulated the genes involved in cell proliferation, apoptosis, cell cycle suppression, and cancer tissue development and differentiation in male Wistar rats. CAP might have a chemopreventive effect against colorectal carcinogenesis [64]. Previously, the effect of chili extracts or pure CAP on colon cancers and HCT116 cells showed induced autophagy [65]. The CAP affects human colorectal cell lines LoVo and SW480 by inducing anti-tumorigenesis, deregulation of β-catenin/TCF-dependent signaling [45], cell death, and increased ROS and pro-apoptotic proteins in cells Colo205 [66]. Recently, the genome profile of Japanase chili pepper was sequenced and determined by Shirasawa et al. [67]. Stimulating food is one of the factor in the development of gastrointestinal tract cancers, with unclear association between chili pepper consumption and the risk of cancer. Chen et al. [68] found that geographic regions influence gastrointestinal cancer risk, particularly in Asian, African, and North American populations, which require greater attention during dietary counseling.
Concluding remarks
Our pilot study pointed out for the first time the inhibitory effect of some chili extracts on Topo I and II activity, as well as the relationship of such extracts to the reduction of metabolic activity and proliferation of human colon cancer cell line HCT116. TMS pepper completely inhibited Topo I and thus is the best candidate for further investigation, suitable for medicinal treatments because their underlying mechanisms differ from those of Topo I inhibitors. In order to confirm the relationship between the inhibition of topoisomerase activity and the antitumor effect of chili extracts, it is necessary to test a wider range of tumor cells as well as the concentration and time scale of the action of such extracts.
Availability of data and materials
All data supporting the findings of this study are available within the paper and within its supplementary materials.
Abbreviations
- HR:
-
Habanero red
- HMR:
-
Habanero maya red
- TMS:
-
Trinidad moruga scorpion
- J:
-
Jalapeno
- SP:
-
Serrano pepper
- HRS:
-
Habanero red savina
- BJ:
-
Bhut Jolokia
- JR:
-
Jamaica rosso
- CAP:
-
capsaicin
- DHK:
-
Dihydrocapsaicin
- pBR:
-
Common plasmid cloning vector in E. coli that is 4361 bp
- Topo I:
-
DNA topoisomerase I
- Topo II:
-
DNA topoisomerase II
- kDNA:
-
Kinetoplast DNA
- MTS:
-
Assay used to assess cell proliferation, cell viability and cytotoxicity
- xCell assay:
-
xCELLigence RTCA systems enable continuous, quantitative, real-time cell analysis
- HCT116:
-
Cell line isolated from the colon of an adult male with colon cancer
- HPLC:
-
High-performance liquid chromatography (HPLC), a technique in analytical chemistry
- RPMI medium:
-
a cell culture medium commonly used to culture mammalian cells
References
Huang CY, Ju DT, Chang CF, Muralidhar RP, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating nonsmall cell lung cancer. Biomed. 2017;7(4):23.
He Q, Shi J. MSN anti-Cancer Nanomedicines: chemotherapy enhancement, overcoming of drug resistance, and metastasis inhibition. Adv Mater (Deerfield Beach, Fla). 2014;26:391–411.
Ahmad R, Ahmad N, Naqvi A, Shehzad A, Al-Ghamdi MS. Role of traditional Islamic and Arabic plants in cancer therapy. J Tradit Complement Med. 2016;7:195–204.
Ijaz S, Akhtar N, Khan MS, et al. Plant derived anticancer agents: a green approach towards skin cancers. Biomed Pharmacother. 2018;103:1643–51.
Seca AML, Pinto DCGA. Plant secondary metabolites as anticancer agents: successes in clinical trials and therapeutic application. Int J Mol Sci. 2018;19:263.
Dong X, Li X, Ding L, Cui F, Tang Z, Liu Z. Stage extraction of capsaicinoids and red pigments from fresh red pepper (Capsicum) fruits with ethanol as solvent. LWT Food Sci Technol. 2014;59(1):396–402.
Stommel RJ, Albrecht E. Genetics. Peppers: botany, production and uses. London: CAB international (ed. V. M. Russo); 2012. 30 p. ISBN-13: 978-1-84593-767-6
Lorencová K. Koření známé i neznámé. GRADA Publishing; 2007. p. 156. ISBN 987-80-247-1934-4
Wang YH, Morris-Natschke SL, Yang J, Niu HM, Long CL, Lee KH. Anticancer principles from medicinal Piper (Hú Jiao) plants. Ż J Tradit Complement Med. 2014;4:8–16.
Aza-Gonzalez C, Núňez-Palenius HG, Neftalí OA. Molecular biology of capsaicinoid biosynthesis in chili pepper. Plant Cell Rep. 2011;30(5):695–706.
Beck J. Capsicum. The genus Capsicum. Edited by Amit Krishna De (Indian science congress association, Calcutta). Taylor & Francis, London. J Nat Prod. 2005;68:964.
Chinn MS, Sharma-Shivappa RR, Cotter JL. Solvent extraction and quantification of Capsaicinoids from Capsicum chinense. Food Bioprod Process. 2011;89:340–5.
Kaiser M, Higuera I, Goycoolea FM (2017) Capsaicinoids: Occurence, chemistry, biosynthesis, and biological effects. Edited by Elhadi M Yahia. Wiley online library. Fruit and vegetable phytochemicals: chemistry and human health, 2nd edition; https://doi.org/10.1002/9781119158042.ch23.
Maihofner C, Heskamp ML. Prospective, non-interventional study on the tolerability and analgesic effectiveness over 12 weeks after a single application of capsaicin 8% cutaneous patch in 1044 patients with peripheral neuropathic pain: first results of the QUEPP study. Curr Med Res Opin. 2013;29:673–83.
Ghiasi Esmaeli ZF, Aghajani Ghazi-Khansari MM, Faramarzi MA, Amani A. Enhancing analgesic and anti-inflammatory effects of capsaicin when loaded into olive oil nanoemulsion: an in vivo study. Int J Pharm. 2019;559:341–7.
Toyoda T, Shi L, Takasu S, Cho YM, Kiriyama Y, Nishikawa A, et al. Anti-inflammatory effects of capsaicin and Piperine on Helicobacter pylori-induced chronic gastritis in Mongolian gerbils. Helicobacter. 2016;21(2):131–42.
Baboota RK, Murtaza N, Jagtap S, Singh DP, Karmase A, Kaur J, et al. Capsaicin-induced transcriptional changes in hypothalamus and alterations in gut microbial count in high fat diet fed mice. J Nutr Biochem. 2014;25:893–902.
Sanati S, Razavi BM, Hosseinzadeh H. A review of the effects of Capsicum annuum L. and its constituent, capsaicin, in metabolic syndrome. Iran J Basic Med Sci. 2018;21:439–48.
Qin SL, Liu SL, Wang RR. Protective effect of capsaicin on against myocardial ischemia-reperfusion injury of rat in vivo. J Sichuan University (Medical Science Edition). 2008;39:550–4.
Park SY, Kim JY, Lee SM, Jun CH, Cho SB, Park CH, et al. Capsaicin induces apoptosis and modulates MAPK signaling in human gastric Cancer cells. Mol Med Rep. 2014;9:499–502.
Siwei CAO, Chen H, **ang S, Hong J, Weng L, Zhu H, et al. Anti-Cancer effects and mechanisms of capsaicin in chili peppers. Am J Plant Sci. 2015;6:3075–81.
Azlan A, Sultana S, Huei CS, Razman MR. (2) Antioxidant, anti-obesity, nutritional and other beneficial effects of different chili pepper. Molecules. 27(3):898. https://doi.org/10.3390/molecules27030898.
Ito K, Nakazato T, Yamato K, Miyakawa Y, Yamada T, Hozumi N, et al. Induction of apoptosis in leukemic cells by homovanillic acid derivative, capsaicin, through oxidative stress: implication of phosphorylation of p53 at Ser-15 residue by reactive oxygen species. Cancer Res. 2004;64:1071–8.
Hail N, Lotan R. Examining the role of mitochondrial respiration in vanilloid-induced apoptosis. J Natl Cancer Inst. 2002;94:1281–92.
Mori A, Lehmann S, O’Kelly J, Kumagai T, Desmond JC, Pervan M, et al. Capsaicin, a component of red peppers, inhibits the growth of androgen-independent, p53 mutant prostate Cancer cells. Cancer Res. 2006;66:3222–9.
Malagarie-Cazenave S, Olea-Herrero N, Vara D, Morell C, Díaz- Laviada I. The vanilloid capsaicin induces IL-6 secretion in prostate PC-3 cancer cells. Cytokine. 2011;54:330–7.
Lee JS, Chang JS, Lee JY, Kim JA. Capsaicin-induced apoptosis and reduced release of reactive oxygen species in MBT-2 murine bladder tumor cells. Arch Pharm Res. 2004;27:1147–53.
Huh HC, Lee SY, Lee SK, Park NH, Han IS. Capsaicin induces apoptosis of cisplatin-resistant stomach cancer cells by causing degradation of cisplatin-inducible aurora-a protein. Nutr Cancer. 2011;63:1095–103.
Lin RJ, Wu IJ, Hong JY, et al. Capsaicin-induced TRIB3 upregulation promotes apoptosis in cancer cells. Cancer Manag Res. 2018;10:4237–48.
Kim YM, Hwang JT, Kwak DW, Lee YK, Park OJ. Involvement of AMPK signaling cascade in capsaicin-induced apoptosis of HT-29 colon cancer cells. Ann N Y Acad Sci. 2007;1095:496–503.
Ip SW, Lan SH, Lu HF, Huang AC, Yang JS, Lin JP, et al. Capsaicin mediates apoptosis in human nasopharyngeal carcinoma npc-tw 039 cells through mitochondrial depolarization and endoplasmic reticulum stress. Human & ExpToxicol. 2012;31:539–49.
Huang SP, Chen JC, Wu CC, Chen CT, Tang NY, Ho YT, et al. Capsaicin-induced apoptosis in human hepatoma HepG2 cells. Anticancer Res. 2009;29:165–74.
Anandakumar P, Kamaraj S, Jagan S, et al. The anticancer role of capsaicin in experimentally induced lung carcinogenesis. Aust J Pharm. 2015;18:19–25.
Friedman JR, Nolan NA, Brown KC, et al. Anticancer activity of natural and synthetic capsaicin analogs. J Pharmacol Exp Ther. 2018;364:462–73.
Clark R, Lee SH. Anticancer properties of capsaicin against human cancer. Anticancer Res. 2016;36(3):837–43.
Arul B, Kothai R. IntechOpen. 2020; https://doi.org/10.5772/intechopen.91897.
Bley Boorman KG, Mohammad B, McKenzie D, Babbar S. A comprehensive review of the carcinogenic and anticarcinogenic potential of capsaicin. Toxicol Pathol. 2012;40:847–73.
Liu RH. Potential synergy of phytochemicals in cancer prevention: mechanism of action. J Nutr. 2004;134(12 Suppl):3479S–85S.
Cheng YY, Hsieh CH, Tsai TH. Concurrent administration of anticancer chemotherapy drug and herbal medicine on the perspective of pharmacokinetics. J Food Drug Anal. 2018;26:S88–95.
Sanchez BG, Bort A, Mateos-Gomez PA, Rodríguez-Henche N, Díaz-Laviada I. Combination of the natural product capsaicin and docetaxel synergistically kills human prostate cancer cells through the metabolic regulator AMP-activated kinase. Cancer Cell Int. 2019;19:54.
Zheng L, Chen J, Ma Z, Liu W, Yang F, Yang Z, et al. Capsaicin enhances anti-proliferation efficacy of pirarubicin via activating TRPV1 and inhibiting PCNA nuclear translocation in 5637 cells. Mol Med Rep. 2016;13(1):881–7.
Nelson VK, Sahoo NK, Sahu M, Sudhan HH, Pullaiah CP, Muraliikrishna KS. In vitro abtcancer activity of Eclipta alba whole plant extract on colon cancer cell HTC-116. BMC Complementary Med Therap. 2020;20:355.
Lee S, Richardson RL, Dashwood RH, Baek SJ. Capsaicin represses transcriptional activity of β-catenin in human colorectal Cancer cells. J Nutr Biochem. 2012;23:646–55.
Lee SH, Clark R. AntiTumorigenic effects of capsaicin in colon cancer. J Food Chem Nanotechnol. 2016;2(4):162–7.
Yang J, Li TZ, Xu GH, Luo BB, Chen YX, Zhang T. Low-concentration capsaicin promotes colorectal Cancer metastasis by triggering ROS production and modulating Akt/mTOR and STAT-3 pathways. Neoplasma. 2013;60:364–72.
Nittis JL. Investigation the biological functions of DNA topisomerases in eukaryotic cells. Biochim Biophys Acta. 1998;1400:63–81.
Wall M, Wani MC, Cooke CE, Palmer KH, McPhail AT, Sim GA. Plant anti-tumor agents I. The isolation and structure of camphotehecin, a novel alkaloid leukemia and antitumor inhibitor from Camptotheca acuminata. Journal of the Americal Society. 1966;88:3888–90.
Delgado JL, Hsieh CM, Chan NL, Hiasa H. Topoisomerase as anticancer targets. Biochem J. 2018;475(2):373–98.
Topcu Z. DNA topisomerases as targets for anticancer drugs. J Clin Pharm Ther. 2021;26:405–16.
Chapa-Oliver AM, Mejía-Teniente L. Capsaicin: from plants to a cancer-suppressing agent. Molecules. 2016;21(8):931.
Garra A, Alkalai-Tuvia S, Telerman A, Paran I, Fallikand E, Elmann (2020) A anti-proliferative activities, phytochemical levels and fruit quality of pepper (Capsicum spp.) following prolonged storage. Int J Food Sci Technol 55: 3574–3584.
Janovec L, Kovacova E, Semelakova M, Kvakova M, Kupka D, Jager D, et al. Synthesis of novel biologically active proflavine ureas designed on the basis of predicted entropy changes. Molecules. 2021;26(16):4860.
Sabolová D, Kristian P, Kožurková M. Proflavine/acriflavine derivatives with versatile biological activities. J Appl Toxicol. 2020;40(1):64–71.
Wessermann K, Markovits J, Jakel C, Capranico G, Kohn KW, Pommier Y. Effects of morpholinyl doxorubicins, doxorubicin, and actinomycin D on mammalian DNA topoisomerases I and II. Mol Pharmacol. 1990;38:38–45.
Kobayashi S, Furukawa M, Dohi C, Hamashima H, Arai T, Tanaka A. Topology effect for DNA structure of cisplatin: topological transformation of cisplatin-closed circular DNA adducts by DNA topoisomerase I. Chem Pharm Bull (Tokyo). 1999;47:783–90.
Okura A, Arakawa H, Oka H, Yoshinair T, Monden Y. Effect of genistein on topoisomerase activity and on the growth of [Al 12] ha-ras-transformed NIH 3T3 cells. Biochem Biophys Res Commun. 1988;157:183–9.
Hsiang YH, Liu LF. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988;48:1722–6.
Thakur A, Singla R, Jaitak V. Coumarins as anticancer agents: a review on synthetic strategies, mechanism of action and SAR studies. Eur J Med Chem. 2015;101:476–95.
Bates SE, Medina-Perez WY, Kohlhagen G, Antony S, Nadjem T, Robey RW, et al. ABCG2 mediates differential resistance to SN-38 (7-ethyl-10-hydroxycamptothecin) and homocamptothecins. Pharmacol Exp Ther. 2004;310:836–42.
Díaz-Laviada I. Effect of capsaicin on prostate cancer cells. Future Oncol Lond Engl. 2010;6:1545–50.
Garufi A, Pistritto G, Cirone M, D’Orazi G. Reactivation of mutant p53 by capsaicin, the major constitutent of peppers. J Exp Clin Cancer Res. 2016;35:136.
Lau JK, Brown KC, Dom AM, Witte TR, Thornhill BA, Crabtree CM, et al. Capsaicin induces apoptosis in human small cell lung cancer via the TRPV6 receptor and the calpain pathway. Apoptosis. 2014;19:1190–201.
Kim HA, Kim MS, Kim SH, Kim YK. Pepper seed extract suppresses invasion and migration of human breast cancer cells. Nutr Cancer. 2014;66(1):159–65.
Caetanoa Ramos BF, Tablasb MB, Ferreira Pereirab NE, de Mourab NA, Carvalhob RF, Marchesan Rodriguesa MA, et al. Capsaicin reduces genotoxicity, colonic cell proliferation and preneoplastic lesions induced by 1,2-dimethylhydrazine in rats. Toxicol Appl Pharmacol. 2018;338:93–102.
Oh SH, Kim YS, Lim SC, Hou YF, Chang IY, You HJ. Dihydrocapsaicin (DHC), a saturated structural analog of capsaicin, induces autophagy in human cancer cells in a catalase regulated manner. Autophagy. 2008;4:1009–19.
Lu HF, Chen YL, Yang JS, Yang YY, Liu JY, Hsu SC, et al. Antitumor activity of capsaicin on human colon cancer cells in vitro and Colo 205 tumor xenografts in vivo. J Agric Food Chem. 2010;58:12999–3005.
Shirasawa K, Hosokawa M, Yasui Y, Toyoda A, Isobe S. Chromosome-scale genome assembly of a Japanese chili pepper landrace, Capsicum annuum “Takanotsume”. DNA Res. 2023;30(1):dsac052. https://doi.org/10.1093/dnares/dsac052.
Chen C, Zhang M, Zheng X, Lang H. Association between chili pepper consumption and risk of gastrointestinal-tract cancers. A meta-analysis. Front Nutr. 2022;9:935865. https://doi.org/10.3389/fnut.2022.935865.
Acknowledgements
The authors are grateful to Gabriela Dye for proofreading the manuscript.
Funding
This study was supported by funds from The Scientific Grant Agency of the Ministry of Education of the Slovak Republic and the Slovak Academy of Sciences (VEGA 1/0037/22, VEGA 1/0073/22).
Author information
Authors and Affiliations
Contributions
M.S. and T.H. proposed the project and M.S. designed experiments. Cultivation of cells was performed by M.S. and Z.T. Cultivation of pepers, extraction and HPLC analysis of CAP and DHK was performed by T.H., P.O. and P.P. Nuclease activity and Topo I, II assays were performed by M.K., K.K. and S.S. MTS analysis and statistical analyses were performed by M.S. xCell analysis were performed by M.S. and Z.G. The manuscript was prepared by M.S., T.H. and M.K. P.S. and PP. consulted progress and approved the final manuscript. The manuscript has been read and approved by all named authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Experimental research on plants complies with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hudáková, T., Šemeláková, M., Očenáš, P. et al. Chili pepper extracts, capsaicin, and dihydrocapsaicin as potential anticancer agents targeting topoisomerases. BMC Complement Med Ther 24, 96 (2024). https://doi.org/10.1186/s12906-024-04394-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-024-04394-5