Abstract
Endometriosis is a common gynecological disease, and its underlying mechanisms remain elusive. Patients are at a higher risk of recurrence after surgery or drug withdrawal. In this study, to identify a potentially effective and safe therapy for endometriosis, we screened potential target genes of kaempferol on endometriosis using network pharmacology and further validation. Network pharmacology showed kaempferol may suppress migratory and invasive properties by modulating the phosphoinositide 3-kinase (PI3K) pathway and its downstream target matrix metalloproteinase (MMP)9. Furthermore, in vitro experiments showed that kaempferol repressed the migration and invasion of endometrial cells, and this effect may be involved in mediating the PI3K-related genes, phosphatase and tensin homolog (PTEN) and MMP9. Network pharmacology and in vitro experiments showed that kaempferol, repressed the implantation of endometrial cells and formation of ectopic lesions by inhibiting migration and invasion and regulating PTEN and MMP9, which may be associated with the PI3K pathway.
Similar content being viewed by others
Introduction
Endometriosis is a common gynecological disease that affects more than 5% of women during the reproductive period [1,2,3]. Regardless of being benign, endometriosis has some malignant features, including migration, invasion, and recurrence. The usual clinical characteristics of endometriosis are dysmenorrhea, which gradually worsens, dyspareunia, irregular menstruation, and infertility. This dramatically affects women’s daily lives. The underlying mechanisms of endometriosis remain unclear. Endometriosis is currently treated with surgery and hormone therapy. However, hormone therapies have some side effects such as anovulation and menstrual irregularities. Furthermore, endometriosis is vulnerable to recurrence (approximately 40%) within five years of drug withdrawal, even after surgery [2, 3]. According to the retrograde menstruation theory [1], the endometrium can invade and implant ectopic tissues other than uterus. Migration and invasion play an essential role in the occurrence and development of ectopic lesions, and drugs that alleviate migration and invasion are urgently required [4]. In this study, we introduced kaempferol in the treatment of endometriosis. Using network pharmacology analysis, we predicted that kaempferol could inhibit the migration and invasion of endometriosis, and verified the efficacy of kaempferol in the treatment of endometriosis through experimental validation in vitro.
Natural products and biomolecules have health-promoting benefits against a much wider range of ailments [5,6,7]. Recently, an increasing number of studies have focused on the anti-tumor effect of kaempferol and it has been widely applied in various cancers, such as pancreatic, stomach, oral, liver, kidney, and brain tumors [8,9,10,11]. Kaempferol suppresses the growth and development of cancer by regulating apoptosis, migration, and invasion via various signaling pathways [8,9,10,11,12,13,14]. For example, kaempferol inhibits migration and invasion by downregulating the extracellular regulating kinase (ERK)1/2 signaling pathway and matrix metalloproteinase-2(MMP2) expression in oral cancers [10]. In hormone-related cancers, such as breast cancer, kaempferol represses the expression of ERα [15]. Endometrium, a hormone-dependent condition, also possesses potential migratory and invasive traits. We hypothesized that kaempferol may be a potential therapeutic agent for endometriosis. We found that kaempferol can modulate integrin binding, ubiquitin, and ubiquitin-like protein ligase binding through the phosphoinositide 3-kinase (PI3K) pathway [16, 17], as predicted by network pharmacology analysis. In this study, we tested whether kaempferol suppressed the migration and invasion of endometriosis and paved the way for therapy with kaempferol in the future. Figure 1 showed the complete flow chart of this study.
Materials and methods
Prediction of targets of kaempferol
The kaempferol was analyzed using the TCMSP database (https://old.tcmsp-e.com/tcmspsearch.php?qs=molecule_name&q=kaempferol&token=d49dd3dbca4e2d3b947698f09744e051) to identify the known drug targets. And the two-dimensional structure of the kaempferol obtained from PubChem was imported into the SwissTargetPrecision database (http://www.swisstargetprediction.ch/), and a threshold (probability > 0.6) was set to obtain a possible target for each combination.
Prediction of endometriosis targets
The keyword “endometriosis” was searched in three different disease gene databases: (1) GeneCards Human Gene database (https://www.genecards.org/Search/Keyword?queryString=endometriosis); (2) DisGeNET database (https://www.disgenet.org/browser/0/1/0/C0014175/); (3) Online Mendelian Inheritance in Man (OMIM) database (https://omim.org/search?index=entry&start=1&limit=10&sort=score+desc%2C+prefix_sort+desc&search=endometriosis). UniProt (http://www.UniProt.org) was used to obtain the gene symbols of all targets (endometriosis and kaempferol), and the information was used for subsequent network pharmacological data analysis. (4) Datasets contain endometriosis tissue and normal endometrial tissue sequencing results from GEO database (http://www.ncbi.nih.gov/geo) to download. The raw data were downloaded as MINiML files. Using the limma package in the R software to study the differentially expressed mRNA. The adjusted P-value was analyzed to correct the false positive results in GEO datasets. “Adjusted P < 0.05 and Log (Fold Change) >1 or Log (Fold Change)<−1” were defined as the threshold for the differential expression of mRNAs.
Screening for key targets
The Venny R package was employed to map the targets of kaempferol and the known therapeutic targets of endometriosis to build a Venn diagram. We combined the crossed targets of kaempferol and endometriosis after deleting the duplicate targets of endometriosis. These cross targets were defined as critical targets for the treatment of endometriosis.
We used the Cytoscape software (version 3.7.2, Boston, MA, USA) to construct the drug-ingredient-target interaction network.
Protein-Protein Interaction (PPI) network construction
The key targets were subjected to PPI analysis using the STRING database (https://string-db.org/), and the species “Homo sapiens” was selected to generate a PPI network. After processing the data by R software (R version 4.1.2), setting the minimum required interaction score highest confidence (0.900) ,we obtained the top 30 hub genes. Subsequently, we imported the hub gene data into the Cy software and obtained the HUB gene PPI network diagram.
Gene Ontology (GO) and Kyoto Encyclopedia of Geld genomes (KEGG) analyses of hub genes
We imported 30 critical genes into the Bioconductor library of R software to perform GO and KEGG pathway enrichment analyses. The screening criteria were set at P<0.05, Q<0.05, and the restricted species was Homo sapiens. GO and KEGG enrichment analyses revealed the top 15 results. Bar and bubble graphs represent the visualization, and a pathway atlas is presented for related pathways that meet clinical significance.
Cell culture
The Ishikawa cells were used in this study. The Ishikawa cell line was obtained from the Basic Medical Laboratory of Qilu Hospital. The cells were cultured in DMEM (Life Technologies, USA) containing 1% penicillin/streptomycin (Gibco, USA) and 10% FBS (Gibco, USA) in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were passaged when they reached 85% confluence and stored in liquid nitrogen for long-term preservation.
Cell viability assay
Kaempferol was purchased from Solarbio Science &Technology Co.,Ltd (Bei**g, China) [18]. The purity of kaempferol used in the following experiments was>98%. Kaempferol was dissolved in DMSO at a concentration of 200 mmol/L.
Ishikawa cells were seeded in 96-well plates at a density of 5,000 cells per well and cultured in a complete cell culture medium for 24 h for recovery. Next, the medium was changed to DMEM with 0, 25, 50, 75, 100, 150, and 200 µmol/L kaempferol, without FBS, and cultured for 48 h. Then, 10 μL of sterile Cell Counting Kit (CCK)8 solution (Bimake, USA) was added to each well and incubated for another 1.5 h at 37 °C. Absorbance was measured at 450 nm using a microplate reader.
Colony-forming assay
Ishikawa cells were seeded at a density of 500 cells per well in a six-well plate and cultured in a complete cell culture medium for 6 h for recovery [19]. Kaempferol was added to the medium at different concentrations, with end densities of 0, 25, 50, and 75 µmol/L. The cells were cultured for a week, and then the medium was gently removed, fixed with 4% paraformaldehyde for 10 min, stained with crystal violet for 15 min, washed, and observed under a microscope.
Wound healing test
Ishikawa cells were seeded at a density of 10, 000 cells per well in a six-well plate and cultured in a complete medium until they reached 85% confluence [20]. Then, a scratch was made to cause a wound, the cells were gently washed, and the remaining cells were incubated with a medium containing DMEM and 0, 25, 50, and 75 µmol/L kaempferol without FBS. The wound surface areas were measured for 24 h to evaluate the ability of the cells to migrate.
Transwell test
The transwell plates were coated with gelatin and cultured in a complete medium for 6 h to recover [21]. Ishikawa cells were diluted with serum-free DMEM containing 0, 25, 50, and 75 µmol/L kaempferol, and inoculated into the upper well of the transwell plate at a density of 100,000 cells per well. The lower well was DMEM complete medium containing 20%FBS to aid their migration. The cells were then incubated in a cell culture container for 24 h. Then, the cells were fixed with 4% paraformaldehyde for 10 min, and the cells in the upper side of the wells were scratched gently; the cells on the lower side were stained with crystal violet and counted under a microscope.
Real-time RT-PCR
Ishikawa cells were seeded at a density of 10,000 cells per well in a six-well plate overnight. Afterward, the cells were incubated with the medium containing DMEM and 0, 25, 50, and 75 µmol/L kaempferol [22]. Total RNA was collected using a RNeasy mini kit (RNA Fast 200, China), according to the manufacturer’s instructions. Then, cDNA was synthesized from 1 μg of total RNA using a SureScript First-Strand cDNA Synthesis Kit (GeneCopoeia, USA). Finally, the expression levels of GAPDH, PTEN, MMP1, and MMP9 were measured using an all-in-one qRT-PCR detection kit (GeneCopoeia, USA). The sequences of the primers were as follows: PTEN, 5′-CAAGATGATGTTTGAAACTATTCCAATG-3′ (sense) and 5′-CCTTTAGCTGGCAGACCACAA-3′ (antisense); MMP1, 5′-CACAAACCCCAAAAGCGTGT-3′ (sense) and 5′-TCGGCAAATTCGTAAGCAGC-3′ (antisense); MMP9, 5′-TCTGCCCCGGACCAAGGATA-3ʹ (sense) and 5′-ACATAGGGTACATGAGCGCC-3′ (antisense); and GADPH, 5′-GCACCGTCAAGGCTGAGAAC-3′ (sense) and 5′-TGGTGAAGACGCCAGTGGA-3′ (antisense). Melting curve analysis was used to confirm the amplification specificity. The quantification data were analyzed using LightCycler analysis software (version 4.0; Roche Applied Science, Mannheim, Germany). Relative expression was normalized to that of GAPDH.
Western blot assay
Ishikawa cells were seeded at a density of 10,000 cells per well in a six-well plate overnight. Afterward, the cells were incubated with the medium containing DMEM and 0, 25, 50, and 75 µmol/L kaempferol for 24 hours. Total proteins were extracted with RIPA buffer (Solarbio, #R0020) which added protease inhibitors (Solarbio, #P0100) and phosphatase inhibitors (Solarbio, #P1260). The lysates were separated on SDS-PAGE gels under 80V. After that, the proteins were immediately trans-blotted to a Millipore PVDF membrane (0.2 µm) under constant current. The blot was blocked with a protein-free rapid blocker(Epizyme,#PS108P) at room temperature for 1 h, and then incubated overnight with primary antibody(abcam, #ab267787;Huabio, # HA721136) at 4℃.Next day, the blots were washed with TBST for 3 times and incubated with secondary antibodies for 1 h. The blots were detected by ECL reagent (Millipore) using a Tanon System for visualization after washing with TBST for 3 times. GAPDH expression was used to quantify the data. ImageJ(2.3.0) was used to analysis (version 5.2.1).
Statistical analysis
SPSS 22.0 was used to analyze the data. Quantitative data are presented as the mean ± SD of at least three independent experiments. One-way ANOVA were employed to compare the means. Differences were considered statistically significant if the p-value was <0.05 or <0.01.
Results
The candidate targets of kaempferol
According to the TCMSP, and SwissTargetPrecision databases, we identified a total of 148 potential targets after deduplication.
Prediction of therapeutic targets of endometriosis
We searched the GeneCards Human Gene Database, DisGeNET database, and Online Mendelian Inheritance in Man database for “endometriosis”. In addition, the differentially expressed genes in endometriosis compared with normal tissues were analyzed from two data sets GSE23339 and GSE25628 in GEO database (Fig. 2 A-E). A total of 1188 targets were identified after deduplication.
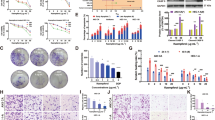
A The boxplot of the normalized data. Different colors represent different datasets. Rows represent samples, and columns represent the gene expression values in the samples. B PCA results before batch removal for multiple datasets. Different colors represent different datasets. As shown in the schematic diagram, three datasets are separated without any intersection. C PCA results after batch removal, as shown in the schematic diagram shows the intersection of three datasets, which can be used in subsequent analysis. D Volcano plot:The volcano plot was constructed using the fold change values and P-adjust. Red dots indicate upregulated genes; blue dots indicate downregulated genes. E The heatmap of the differential gene expression, different colors represent the trend of gene expression in different tissues. The top 50 up-regulated genes and top 50 down-regulated genes were showed in this figure. F Venn diagram of targets of active ingredients of kaempferol and those related to endometriosis
Screening for key targets
Using the Venny R package, we obtained 87 key targets to establish a Venn diagram (Fig. 2F).
Construction of the PPI network and selection of HUB genes
As depicted in Fig. 3, we obtained the PPI network using the STRING database and the restricted species Homo sapiens. We obtained 30 hub genes through PPI results and the Cytoscape plug-in cytoHubba (Fig. 4).
GO and KEGG pathway enrichment analyses
We imported 30 critical genes into the Bioconductor library of the R software for GO and KEGG pathway enrichment analyses. The top 15 results and bar and bubble graphs were obtained.
GO analysis revealed that mitogen-activated protein kinase (MAPK) activity, R-(mothers against decapentaplegic) SMAD binding, integrin binding, ubiquitin, and ubiquitin-like protein ligase binding may be vital biological processes related to the treatment of endometriosis by kudingcha. KEGG analysis showed that the MAPK, PI3K-Akt, Forkhead box O (FoxO), and HIF-1 signaling pathways might be the central pathways associated with the treatment of endometriosis by kaempferol (Fig. 5).
Kaempferol impairs cell viability and proliferation of Ishikawa cells in a concentration-dependent pattern
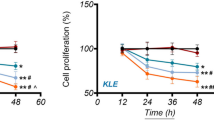
To evaluate the effect of kaempferol on cell viability, Ishikawa cells were treated with different concentrations of kaempferol for 48 h. Then, the CCK8 assay was used to assess cell viability. The results showed that at low concentrations of 25, 50, and 75 µmol/L, kaempferol did not affect the viability of Ishikawa cells (P>0.05); however, at high concentrations of 100, 150, and 200 µmol/L, kaempferol decreased the viability of Ishikawa cells (P<0.01) (Fig. 6A). The effect of kaempferol on the proliferation of Ishikawa cells was evaluated via a colony-forming assay. The results showed no significant difference in colony numbers between 0, and 25 µmol/L kaempferol, whereas the number of cell colonies decreased significantly at 50, and 75 µmol/L kaempferol compared to 0 µmol/L (P<0.01) (Fig. 6B and C).
Effect of kaempferol on the viability and proliferation of Ishikawa cells. A CCK8 assay showed that kaempferol did not affect the viability of Ishikawa cells at low concentrations (25, 50, and 75 µmol/L) while reduced the viability of endo cells in a concentration-dependent pattern at high concentrations (100, 150, and 200 µmol/L). B and (C) Colony-forming assay showed that kaempferol did not affect the proliferation of Ishikawa cells at 25 µmol/L but had an inhibitory effect at 50 and 75 µmol/L. ** P<0.01, ***P<0.001, **** P<0.0001
Kaempferol decreases cell migration and invasion of Ishikawa cells
A wound healing assay was performed to evaluate the effect of kaempferol on the migration of Ishikawa cells. The results showed no significant difference in the migration distance between the 0 and 25 µmol/L kaempferol treatment groups (P>0.05); however, the migration distance was significantly decreased by 50 and 75 µmol/L kaempferol (P<0.05, or P<0.01) (Fig. 7A and B). Cell invasion ability was evaluated via a Transwell assay. The results showed no significant difference in the number of cells on the lower surface between the 0 and 25 µmol/L kaempferol groups; in contrast, the numbers were significantly decreased in the 50 and 75 µmol/L kaempferol-treated groups compared to the control group (P<0.01) (Fig. 7C and D).
Effect of kaempferol on the migration and invasion of Ishikawa cells. A and B show that kaempferol inhibits the migration of Ishikawa cells in a wound healing assay; C and D show that kaempferol impairs the invasion ability of Ishikawa cells in a transwell assay. ** P<0.01, ***P<0.001, **** P<0.0001
Kaempferol upregulated the expression of PTEN while downregulated that of MMP9 in Ishikawa cells
Real-time RT-PCR was performed to determine the potential mechanism of action of kaempferol in endometriosis. The results demonstrated that expression of PTEN was attenuated after treatment with 50 and 75 µmol/L kaempferol (Fig. 8A). At the same time, 25 µmol/L kaempferol treatment did not result in differential alterations compared with the control group. In addition, MMP9 expression was significantly inhibited after treatment with 25, 50, and 75 µmol/L kaempferol (Fig. 8C). However, there was no significant difference in MMP1 expression after treatment with kaempferol (Fig. 8B). We further performed Western blot assay on the changes of PTEN. The results showed that the protein expression level of PTEN increased with kaempferol concentration (Fig. 8D).
Effect of kaempferol on the predicted genes associated with migration and invasion of Ishikawa cells. A Kaempferol significantly upregulated the expression of PTEN in endometrial cells; B Kaempferol did not significantly affect MMP1 levels in endometrial cells; C Kaempferol significantly reduced MMP9 levels in endometrial cells; D Kaempferol enhanced the expression of PTEN in endometrial cells by western blotting. *P<0.05, ** P<0.01, ***P<0.001, **** P<0.0001
Discussion
Traditional Chinese medicine has been used to treat various diseases for thousands of years and has achieved considerable accomplishments. However, the mechanisms remain ambiguous because herbs exert multifaceted effects through multiple pathways. The main ingredients and target genes can be identified by network analysis. In this study, we aimed to identify the primary target genes of kaempferol involved in the treatment of endometriosis. Using GO analysis, we found that MAP kinase activity, R-SMAD binding, integrin binding, ubiquitin, and ubiquitin-like protein ligase binding may be vital biological processes affected by kaempferol. In addition, KEGG analysis revealed that the MAPK, PI3K-Akt, FoxO, and Hypoxia-inducible factors (HIF)-1 signaling pathways may be the central signaling pathways associated with the treatment of endometriosis by kaempferol. Subsequently, we assessed the migration and invasion of endometrial cells. In addition, the expression of PTEN and MMPs, which are target genes associated with the PI3K/Akt/MMP signaling pathway, was evaluated by real-time PCR, considering the data mining results. The experiments suggested that kaempferol, may inhibit the migration and invasion of endometrial cells by regulating the PI3K/Akt/MMP signaling pathway, coinciding with predictions by network pharmacology and data mining.
Endometriosis is a common gynecological disease. Because of the migration and invasion of epithelial cells, the endometrium can develop ectopic lesions in organs other than the uterus. Kaempferol has been shown to suppress migration and invasion in many cancers [5, 13, 14, 23]. For instance, kaempferol inhibits the migration and invasion of human retinal pigment epithelial cells [24]. Recent studies have shown that kaempferol represses migration and invasion via various signaling pathways. Kaempferol regulates PI3K/ mammalian target of rapamycin (mTOR)/MMPs protein pathways in liver cancers [25]. Moreover, kaempferol blocks the MAPK signaling pathway and downstream MMP expression to influence migration and invasion in rheumatoid arthritis [26]. In this study, we used the Cytoscape software to identify the main target of kaempferol via drug-ingredient-target interaction network and GO and KEGG analyses. The results demonstrated that kaempferol might alleviate migration and invasion in endometriosis via the PI3K signaling pathway and downstream MMP expression. The subsequent experiments involving wound healing and transwell assays showed that kaempferol reduced migration and invasion without affecting cell proliferation, which was in accordance with the prediction. Furthermore, RT-PCR was used to quantify the genes associated with the signaling pathways. The results showed that kaempferol increased the expression of PTENs, which are related to the PI3K pathway, and decreased the expression of MMP9.
The PI3K/AKT/mTOR pathway participates in various processes including proliferation, migration, invasion, and apoptosis. Several studies have shown that the inactivation of this pathway could prevent the recurrence of endometriosis [27]. And the results of KEGG analysis also identified that the effect of kaempferol on endometriosis may be related to PI3K/AKT/mTOR pathway [28,29,30]. The downstream gene, PTEN, is downregulated in both eutopic and ectopic endometrium from patients with endometriosis [31, 32]. Additionally, PTEN and platelet-derived growth factor (PDGF) play a role in the migration and invasion of endothelial cells and enhance the formation of ectopic lesions [33]. Migration and invasion are vital processes in the development and metastasis of cancer [34, 35]. The MMPs are central to many pathophysiological activities such as apoptosis, migration, and growth [36]. Among MMPs, MMP2 has been shown to mediate migration aninvasion in gliomas through focal adhesion kinase (FAK) and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling [37, 38]. Additionally, MMP1 and MMP14 are associated with migration and invasion [34, 35]. As far as endometriosis is concerned, increased expression of MMPs has been used as a biomarker in endometriosis [39]. Additionally, the PI3K pathway and MMP9 participate in modulating the migration and invasion of endometrial cells [40]. In this study, the results of wound healing and transwell assays showed that kaempferol inhibited the migration and invasion of endometrial cells. Kaempferol attenuated the expression of PTEN and led to downregulation of MMP9, as suggested by RT-PCR results. These results indicated that kaempferol suppressed the migration and invasion of endothelial cells by modulating the expression of genes associated with the PI3K pathway.
However, this study has several limitations. First, the analysis was performed based on TCMSP, ETCM, and SwissTargetPrecision databases. This might have led to biased results. Second, data mining analysis is a prediction and requires further validation. We validated the selected genes and pathways in addition to this research. This may be omitted.
In conclusion, kaempferol plays vital roles in the migration and invasion of endometriosis, as indicated by network pharmacology and data mining. Experiments validated the protective effect of kaempferol, and the potential impact that may be involved in the regulation of PI3K/MMPs.
Availability of data and materials
The data in the current study come from TCMSP database (https://old.tcmsp-e.com/tcmsp.php) and STRING database (https://string-db.org/). All data generated or analyzed in this study are included in this article and its additional files.
Abbreviations
- PI3K:
-
Phosphoinositide 3-kinase
- MMP:
-
Matrix metalloproteinase
- PTEN:
-
Phosphatase and tensin homolog
- ERK1/2:
-
Extracellular regulating kinase1/2
- TCMSP:
-
Chinese Medicine Systems Pharmacology
- ETCM:
-
Encyclopedia of Traditional Chinese Medicine
- OB:
-
Oral bioavailability
- DL:
-
Drug-likeness
- OMIM:
-
Online Mendelian Inheritance in Man
- PPI:
-
Protein-protein interaction
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Geld genomes
- CCK:
-
Cell Counting Kit
- MAPK:
-
Mitogen-activated protein kinase activity
- R-SMAD:
-
R-mothers against decapentaplegic
- FoxO:
-
Forkhead box O
- HIF-1:
-
Hypoxia-inducible factors-1
- mTOR:
-
mammalian target of rapamycin
- PDGF:
-
Platelet-derived growth factor
- FAK:
-
Focal adhesion kinase
- JAK:
-
Janus kinase
- STAT:
-
Signal transducer and activator of transcription
References
Li J, Ma J, Fei X, Zhang T, Zhou J, Lin J. Roles of cell migration and invasion mediated by twist in endometriosis. J Obstet Gynaecol Res. 2019;45:1488–96.
Liu H, Zhang W, Wang L, Zhang Z, **ong W, Zhang L, et al. GLI1 is increased in ovarian endometriosis and regulates migration, invasion and proliferation of human endometrial stromal cells in endometriosis. Ann Transl Med. 2019;7:663–663.
Lu Q, Huang Y, Wu J, Guan Y, Du M, Wang F, et al. T-cadherin inhibits invasion and migration of endometrial stromal cells in endometriosis. Hum Reprod. 2020;35:145–56.
Mc Cormack B, Maenhoudt N, Fincke V, Stejskalova A, Greve B, Kiesel L, et al. The ellagic acid metabolites urolithin A and B differentially affect growth, adhesion, motility, and invasion of endometriotic cells in vitro. Hum Reprod. 2021;36:1501–19.
Mu C, Sheng Y, Wang Q, Amin A, Li X, **e Y. Potential compound from herbal food of Rhizoma Polygonati for treatment of COVID-19 analyzed by network pharmacology: Viral and cancer signaling mechanisms. J Func Foods. 2021;77:104149.
Al-Shamsi M, Amin A, Adeghate E. Effect of vitamin C on liver and kidney functions in normal and diabetic rats. Ann N Y Acad Scie. 2006;1084:371–90.
Murali C, Mudgil P, Gan CY, Tarazi H, El-Awady R, Abdalla Y, Amin A, Maqsood S. Camel whey protein hydrolysates induced G2/M cellcycle arrest in human colorectal carcinoma. Scie Rep. 2021;11:62.
Hung TW, Chen PN, Wu HC, Wu SW, Tsai PY, Hsieh YS, et al. Kaempferol inhibits the invasion and migration of renal cancer cells through the downregulation of AKT and FAK Pathways. Int J Med Sci. 2017;14:984–93.
Lee J, Kim JH. Kaempferol Inhibits Pancreatic Cancer Cell Growth and Migration through the Blockade of EGFR-Related Pathway In Vitro. PLoS One. 2016;11:e0155264.
Lin CW, Chen PN, Chen MK, Yang WE, Tang CH, Yang SF, et al. Kaempferol reduces matrix metalloproteinase-2 expression by down-regulating ERK1/2 and the activator protein-1 signaling pathways in oral cancer cells. PLoS One. 2013;8:e80883.
Zhu G, Liu X, Li H, Yan Y, Hong X, Lin Z. Kaempferol inhibits proliferation, migration, and invasion of liver cancer HepG2 cells by down-regulation of microRNA-21. Int J Immunopathol Pharmacol. 2018;32:2058738418814341.
Liu Z, Yao X, Sun B, Jiang W, Liao C, Dai X, et al. Pretreatment with kaempferol attenuates microglia-mediate neuroinflammation by inhibiting MAPKs-NF-κB signaling pathway and pyroptosis after secondary spinal cord injury. Free Radic Biol Med. 2021;168:142–54.
Chen HJ, Lin CM, Lee CY, Shih NC, Peng SF, Tsuzuki M, et al. Kaempferol suppresses cell metastasis via inhibition of the ERK-p38-JNK and AP-1 signaling pathways in U-2 OS human osteosarcoma cells. Oncol Rep. 2013;30:925–32.
Li C, Zhao Y, Yang D, Yu Y, Guo H, Zhao Z, et al. Inhibitory effects of kaempferol on the invasion of human breast carcinoma cells by downregulating the expression and activity of matrix metalloproteinase-9. Biochem Cell Biol. 2015;93:16–27.
Hung H. Inhibition of estrogen receptor alpha expression and function in MCF-7 cells by kaempferol. J Cell Physiol. 2004;198:197–208.
Ma T, Liu P, Wei J, Zhao M, Yao X, Luo X, et al. Imperatorin alleviated endometriosis by inhibiting the activation of PI3K/Akt/NF-κB pathway in rats. Life Sci. 2021;274:119291.
Hung SW, Zhang R, Tan Z, Chung JPW, Zhang T, Wang CC. Pharmaceuticals targeting signaling pathways of endometriosis as potential new medical treatment: a review. Med Res Rev. 2021;41:2489–564.
Keay KA, Argueta MA, Zafir DN, Wyllie PM, Michael GJ, Boorman DC. Evidence that increased cholecystokinin (CCK) in the periaqueductal gray (PAG) facilitates changes in Resident-Intruder social interactions triggered by peripheral nerve injury. J Neurochem. 2021;158:1151–71.
Youness RA, Gad AZ, Sanber K, Ahn YJ, Lee GJ, Khallaf E, Hafez HM, Motaal AA. Targeting hydrogen sulphide signaling in breast cancer. Journal of advanced research. 2022;27:177–90.
Zhang F, Ni ZJ, Ye L, Zhang YY, Thakur K, Cespedes-Acuña CL. Asparanin A inhibits cell migration and invasion in human endometrial cancer via Ras/ERK/MAPK pathway. Food Chem Toxicol. 2021;150:112036.
Wang M, Zhang G, Zhang Y, Cui X, Wang S, Gao S, et al. Fibrinogen alpha chain knockout promotes tumor growth and metastasis through integrin-AKT signaling pathway in lung cancer. Mol Cancer Res. 2020;18:943–54.
Zhang X, Kan H, Liu Y, Ding W. Plumbagin induces Ishikawa cell cycle arrest, autophagy, and apoptosis via the PI3K/Akt signaling pathway in endometrial cancer. Food Chem Toxicol. 2021;148:111957.
Li S, Yan T, Deng R, Jiang X, **ong H, Wang Y, et al. Low dose of kaempferol suppresses the migration and invasion of triple-negative breast cancer cells by downregulating the activities of RhoA and Rac1. Onco Targets Ther. 2017;10:4809–19.
Chien HW, Wang K, Chang YY, Hsieh YH, Yu NY, Yang SF, et al. Kaempferol suppresses cell migration through the activation of the ERK signaling pathways in ARPE-19 cells. Environ Toxicol. 2019;34:312–8.
Yang G, **ng J, Aikemu B, Sun J, Zheng M. Kaempferol exhibits a synergistic effect with doxorubicin to inhibit proliferation, migration, and invasion of liver cancer. Oncol Rep. 2021;45:32.
Pan D, Li N, Liu Y, Xu Q, Liu Q, You Y, et al. Kaempferol inhibits the migration and invasion of rheumatoid arthritis fibroblast-like synoviocytes by blocking activation of the MAPK pathway. Int Immunopharmacol. 2018;55:174–82.
Barra F, Ferro Desideri L, Ferrero S. Inhibition of PI3K/AKT/mTOR pathway for the treatment of endometriosis. Br J Pharmacol. 2018;175:3626–7.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–51.
Madanes D, Bilotas MA, Bastón JI, Singla JJ, Meresman GF, Barañao RI, et al. PI3K/AKT pathway is altered in the endometriosis patient’s endometrium and presents differences according to severity stage. Gynecol Endocrinol. 2020;36:436–40.
Takeuchi A, Koga K, Satake E, Makabe T, Taguchi A, Miyashita M, et al. Endometriosis triggers excessive activation of primordial follicles via PI3K-PTEN-Akt-Foxo3 Pathway. J Clin Endocrinol Metab. 2019;104:5547–54.
Lim Y, Lee M, Jeong H, Kim H. Involvement of PI3K and MMP1 in PDGF-induced migration of human adipose-derived stem cells. Dev Reprod. 2017;21:167–80.
Chen N, Zhang G, Fu J, Wu Q. Matrix metalloproteinase-14 (MMP-14) downregulation inhibits esophageal squamous cell carcinoma cell migration, invasion, and proliferation. Thorac cancer. 2020;11:3168–74.
Li Y, Huang H, Ye X, Huang Z, Chen X, Wu F, et al. miR-202-3p negatively regulates MMP-1 to inhibit the proliferation, migration and invasion of lung adenocarcinoma cells. Cell Cycle. 2021;20:406–16.
Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF. The roles of matrix Metalloproteinases and their inhibitors in human diseases. Int J Mol Scien. 2020;21:9739.
Cho HJ, Park JH, Nam JH, Chang YC, Park B, Hoe HS. Ascochlorin suppresses MMP-2-mediated migration and invasion by targeting FAK and JAK-STAT signaling cascades. J Cell Biochem. 2018;119:300–13.
Zhao H, **ng F, Yuan J, Li Z, Zhang W. Sevoflurane inhibits migration and invasion of glioma cells via regulating miR-34a-5p/MMP-2 axis. Life Sci. 2020;256:117897.
May KE, Villar J, Kirtley S, Kennedy SH, Becker CM. Endometrial alterations in endometriosis: a systematic review of putative biomarkers. Hum Reprod Update. 2011;17:637–53.
Riemma G, Laganà AS, Schiattarella A, Garzon S, Cobellis L, Autiero R, et al. Ion channels in the pathogenesis of endometriosis: a cutting-edge point of view. Int J Mol Sci. 2020;21:1114.
Acknowledgment
We would like to thank Editage (http://www.editage.com) for editing and reviewing this manuscript for English language.
Funding
This work was supported by Shandong Provincial Natural Science Foundation [ZR2021MH113].
Author information
Authors and Affiliations
Contributions
ZXD, LSL, and LJX contributed to designing the study. ZJD organized the database. WJT and ZJD conducted the experimental evaluation. LPF performed the statistical analysis. WJT and ZXD wrote the first draft of the manuscript. ZXD, WJT, and ZJD wrote sections of the manuscript. All authors contributed to the manuscript revision and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
I confirm that all experiments were carried out in accordance with Declaration of Helsinki. And this study did not require the approval from the ETHICS Committee because the in vitro experiments were conducted on cell lines.
Consent for publication
Not applicable.
Competing interests
All the authors declared that they had no conflicts to do the work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, J., Wang, J., Liu, J. et al. Effect and mechanisms of kaempferol against endometriosis based on network pharmacology and in vitro experiments. BMC Complement Med Ther 22, 254 (2022). https://doi.org/10.1186/s12906-022-03729-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-022-03729-4