Abstract
Background
Otosclerosis (OTSC) is among the most common causes of a late-onset hearing loss in adults and is characterized by an abnormal bone growth in the otic capsule. Alteration in the osteoprotegerin (OPG) expression has been suggested in the implication of OTSC pathogenesis.
Methods
A case-control association study of rs2228568, rs7844539, rs3102734 and rs2073618 single nucleotide polymorphisms (SNPs) in the OPG gene was performed in a Tunisian-North African population composed of 183 unrelated OTSC patients and 177 healthy subjects. In addition, a multilocus association and a meta-analysis of existing studies were conducted.
Results
Rs3102734 (p = 0.013) and rs2073618 (p = 0.007) were significantly associated with OTSC, which were predominantly detected in females after multiple corrections. Among the OPG studied SNPs, the haplotypes A-A-C-G (p = 0.0001) and A-A-C-C (p = 0.0004) were significantly associated with OTSC in females. Multilocus association revealed that the SNPs: rs2073618 in OPG, rs1800472 in TGFβ1, rs39335, rs39350 and rs39374 in RELN, and rs494252 in chromosome 11 showed significant OTSC-associated alleles in Tunisian individuals. In addition, meta-analysis of the rs2073618 SNP in Tunisian, Indian and Italian populations revealed evidence of an association with OTSC (OR of 0.826, 95% CI [0.691–0.987], p = 0.035).
Conclusions
Our findings suggest that rs3102734 and rs2073618 variants are associated with OTSC in North African ethnic Tunisian population. Meta-analysis of the rs2073618 in three different ethnic population groups indicated an association with OTSC.
Similar content being viewed by others
Background
Hearing loss (HL) in humans significantly reduces the quality of life and often leads to social isolation. One of the associated causes of acquired hearing impairment is otosclerosis (OTSC). OTSC is characterized by late-onset progressive sensorineural, conductive or mixed HL. The onset of this disease appears principally in the third decade, while the hot spot age is in the sixth decade [1].
OTSC is a bone-related disorder affecting the otic capsule of the middle ear, which in normal cases undergoes very little remodeling after development and ossification of the tissue. The exact mechanism that controls bone metabolism in the otic capsule and turnover within the auditory structures remains largely unknown. Bone is a dynamic tissue controlled by various biochemical, biomechanical and hormonal stimuli. Bone homeostasis is coordinated at the cellular level by a balance between resorption mediated by the osteoclasts and formation mediated by the osteoblasts [2,3,4]. An imbalance between both processes occurs under certain pathological conditions that affect the skeleton, which leads to the development of bone diseases [5].
The etiology of OTSC remains complex. It is a multifactorial disease caused by both environmental factors such as a viral infection and genetic factors. Evidence supporting the genetic role includes familial cases and a twin study revealing the high heritability [6]. Earlier studies suggested the autosomal dominant inheritance with reduced penetrance in OTSC [7]. Different populations-based case-control studies have associated a number of single nucleotide polymorphisms (SNPs) in candidate genes with OTSC such as COL1A1, TGFβ1, BMP2, BMP4, AGT, ACE and FGF2 [8, 9]. In addition, a genome-wide association study identified the region on chr11q13.1 and 7q22.1 in intron 1 to 4 of the RELN gene to be associated with OTSC [10]. Recent studies using massive parallel sequencing in OTSC families identified six rare heterozygous SERPINF1 variants of which three are missense mutations predicted to be deleterious to the protein function [11], and a rare segregating heterozygous frameshift variant in the MEPE gene [12].
Animal models and genetically altered mice studies during the past 20 years have greatly increased our knowledge of the factors that regulate the activity and formation of osteoclasts. In particular, the discovery of the receptor activator of nuclear-κB ligand (RANKL)/RANK/osteoprotegerin (OPG) signaling axis has improved our understanding of the role played by osteoblasts in these processes [13]. OPG, also known as osteoclastogenesis-inhibiting factor, is a cytokine receptor and a member of the tumor necrosis factor (TNF) receptor superfamily encoded by the TNF receptor superfamily member 11B (TNFRSF11B) gene. The OPG protein is implicated in different signal transduction-interfering biological responses, including apoptosis, cytotoxicity, differentiation and proliferation. It interacts with two common TNF family ligands, TNF-related apoptosis-inducing ligand (TRAIL) and RANKL [14, 15]. The soluble receptor OPG acts as an endogenous decoy receptor towards RANKL (Fig. 1). OPG binds RANKL, thus preventing the interaction of RANKL with, and stimulation of RANK [13, 16]. Therefore, OPG inhibits osteoclast differentiation and maturation and induces the apoptosis of activated osteoclasts as demonstrated in vitro [17] and in vivo [18].
Schematic representation of the cellular and molecular players involved in the RANKL/RANK/OPG signaling axis in bone remodeling. The RANKL from the osteoblast-lineage cells binds to the RANK receptor on osteoclast precursor cells and promotes osteoclasts activation and differentiation, resulting in increased bone resorption. Whereas the OPG decoy receptor binds to RANKL leading to inhibition of osteoclastogenesis. OPG also functions as a decoy receptor for TRAIL, and OPG decreased production mediates the TRAIL-induced apoptosis of osteoblasts leading therefore to bone loss
A deficiency in the OPG/RANKL composition induces a range of skeletal diseases such as osteoporosis and bone metastases [19]. Thus, the concentration of RANKL and OPG in bone is a major determinant of bone mass [23, 24].
The analysis of SNPs was performed using different allelic discrimination genoty** assays. All polymorphisms of the OPG gene were amplified from genomic DNA samples using selected primers (Table 1). For both SNPs, rs7844539 (c.817 + 8A > C) and rs2228568 (c.768A > G), Sanger sequencing was used to detect the variations. For rs3102734 (c.30 + 15C > T), polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was performed using HaeIII restriction enzyme. The rs2073618 (c.9C > G) was genotyped using allele-specific polymerase chain reaction [27].
Association analysis of the OPG
For the four studied SNPs (rs2228568, rs7844539, rs3102734 and rs2073618) in the OPG gene, allele and genotype frequencies were calculated using the direct counting method. The Hardy-Weinberg Equilibrium (HWE) for determination of genotype frequency in control groups was examined using the HWE calculator as previously described [28]. Statistical power calculation was determined in case-control groups with significance levels set at 5% using a web browser program (http://osse.bii.a-star.edu.sg/). Statistical analysis of association characteristics of the study cohort was performed using SPSS (SPSS version 17.0 for Windows, Chicago, IL). A p-value (p) of 0.05 was considered significant. Multivariate logistic regression analysis was undertaken to evaluate associations between genotypes and gender of the analyzed OPG gene polymorphisms. Statistical evaluation of the level of odds ratio (OR) and 95% confidence intervals (CI) for the allelic models were calculated using the 2BY2 program by Fisher’s exact test [29] to evaluate the risk of OPG polymorphisms and OTSC (p < 0.05).
Association analysis in OPG, TGFβ1, RELN and chromosome 11
The multilocus association with OTSC was performed using SNPAlyze V8 Pro (Dynacom, Chiba, Japan) for specific polymorphisms reported in the same Tunisian population: rs2073618 (chr8:119964052, C > G) in the OPG gene of the present study, rs1800472 (chr19:41847860, C > T) in the TGFβ1 gene [25], rs39335 (chr7:103453569, A > G), rs39350 (chr7:103467267, C > T) and rs39374 (chr7:103476667, A > G) in the RELN gene, and rs494252 (chr11:64600002, G > T) in chromosome 11 [26]. Allelic combination analysis among OTSC case and control groups, were evaluated by the maximum-likelihood method using the expectation-maximization (EM) algorithm. All statistical analyses were two-tailed with the statistical significance level fixed at p < 0.05. Permutation p-values were calculated by comparing combination frequencies among case and control groups based on 10,000 replications. Only significant combinations that were males, females and both sex-based analysis with a frequency within [0, 10− 4] in case or control groups were considered.
Pairwise linkage disequilibrium (LD) analysis of |D’| and r2 coefficients was assessed for the six variants (rs2073618, rs1800472, rs39335, rs39350, rs39374, rs494252) using SNPAlyze V8 Pro (Dynacom, Chiba, Japan). This analysis was estimated according to HWE model.
Meta-analysis of rs2073618
A meta-analysis was performed for the rs2073618 (c.9C > G) SNP in the OPG gene in order to investigate its genetic effect in previously studied OTSC populations (Tunisian: the current, Indian [24], and Italian [23] populations). The analysis was performed following the fixed-effect model or the random-effects [30] using comprehensive Meta-Analysis Software V2 (Biostat Inc, Englewood, USA). The evaluation of heterogeneity of inter-study variations was performed with Cochran’s Q-test, as a simple Chi-square test [31]. The null hypothesis was that all studies were evaluating the same effect while the rejection of the null hypothesis (p < 0.05) would reveal heterogeneity between studies. Egger’s regression and sensitivity analysis were estimated. In addition, an indicator of heterogeneity (I2) was evaluated to measure the level of inconsistency across studies [32,33,34,35].
Results
Association of OPG SNPs with OTSC
We evaluated the association of previously documented OPG SNPs (rs2228568, rs7844539, rs3102734 and rs2073618, Fig. 2) with OTSC in a Tunisian population. HWE analysis revealed no deviations (p > 0.001) of the genotype frequencies in the control group for each SNP. Statistical power calculations estimated that at a 5% significance level, the population study had sufficient power to detect SNPs effects with rs3102734 and rs2073618 at 48.5 and 51.1%, respectively, while rs2228568 and rs7844539 had limited power to detect an effect at a significance level of 4.7%.
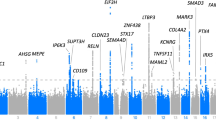
Schematic genomic structure of the OPG gene with the positions of the studied polymorphisms. The filled boxes indicate the protein-coding regions and the white boxes indicate the untranslated regions. The variants (rs2228568, rs7844539, rs3102734 and rs2073618) locations within the OPG gene are marked with arrows. The sizes of exons/introns and positions of the variants within the OPG gene are proportional to the original gene size according to the USC Genome Browser (GRCh37/hg19)
Case-control association analyses revealed no statistically significant genetic differences between patients with OTSC and controls for the polymorphisms rs2228568 (p = 0.483) and rs7844539 (p = 0.483) within exon 4 of the OPG gene. Statistically significant associations were detected between the rs3102734 (p = 0.013) and rs2073618 (p = 0.007), located within exon 1 of the OPG gene, and patients with OTSC compared to healthy controls. Significant allelic associations between the rs3102734 and rs2073618 variants, and OTSC were detected with p = 0.014 and p = 0.0041, respectively (Table 2). The minor allele frequencies (MAFs) in the patients with OTSC accounted for 0.041 in rs3102734, and 0.71 in rs2073618, while the respective MAFs in the healthy controls was 0.09 and 0.619, respectively. In addition, a female gender association with OTSC was detected in rs3102734 and rs2073618 variants (p = 0.046, p = 0.002) but not in males (p = 0.096, p = 0.416), whereas no gender effect was obtained for both rs2228568 and rs7844539 SNPs (Table 3).
OPG haplotypes association with OTSC and linkage disequilibrium analysis
In order to evaluate the potential effects of allelic combinations on the risk of OTSC, haplotype analysis was performed for the four described OPG SNPs [rs2228568 (A > G), rs7844539 (A > C), rs3102734 (C > T), and rs2073618 (C > G)] in the Tunisian population. Five common haplotypes resulted within the OPG gene in OTSC cases and controls, with two being significant (A-A-C-G, p = 0.0135 and A-A-C-C, p = 0.0209, Table 4). Similar results to the genotype analysis, sex-stratified haplotypes analysis revealed gender-specific association of female groups with OTSC for the rs2073618 (c.9C > G). The haplotype A-A-C-G with “G” allele was significantly increased (p = 0.0001) in cases suggested an increased risk of OTSC in females while the A-A-C-C with “C” allele was significantly increased (p = 0.0004) in controls suggested a reduced risk of OTSC in females. The analysis of the haplotype A-A-C-G revealed that the disease-associated “G” allele in the Tunisian population is in line with the previously reported Indian study but with male-specific association.
Multilocus association with OTSC
Allele combinations were examined in order to evaluate the multilocus association effect of selected SNPs with the risk of OTSC. Therefore, the rs2073618 (C > G) SNP in the OPG gene was considered, and previously reported SNPs, located in three different regions rs1800472 (C > T) in the TGFβ1 gene [25], rs39335 (A > G), rs39350 (C > T) and rs39374 (A > G) in the RELN gene and rs494252 (G > T) in the chromosome 11 [26]. Statistical analysis was carried out for the six SNPs (Table 5) and resulted in four common allele combinations detected in otosclerotic and control patients (Table 6). Among these, two allele combinations resulted in significant differences between OTSC cases and controls with a sex-specific association: C-C-A-C-A-G (p = 0.0008) for females and C-C-A-T-G-G (p = 0.0094) for males.
Furthermore, pairwise linkage disequilibrium was calculated according to D’ and r2 statistics for all possible two-way comparisons between the six following SNPs: rs2073618 (C > G) in the OPG gene, rs1800472 (C > T) in the TGFβ1 gene, rs39335 (A > G), rs39350 (C > T) and rs39374 (A > G) in the RELN gene and rs494252 (G > T) in chromosome 11. The degree of LD between these SNPs resulted in strong linkage disequilibrium (D’ > 0.8) for different two-way combinations. The rs1800472 SNP was in complete LD with the three SNPs (rs2073618, rs39350 and rs39374), but also the rs494252 was in complete LD with rs39350 and rs39374 SNPs (Table 7).
Meta-analysis of the association of rs2073618 SNP in the OPG gene with risk of OTSC
A meta-analysis of the rs2073618 (c.9C > G) SNP in the OPG gene was conducted in Tunisian (the present study), Indian and Italian populations which included a total of 528 OTSC and 511 control samples. Statistical significance was evaluated through Z and p-value. The resulted association outcomes were in line with the previously reported in Indian population [24]. A random effects model was selected with a significant heterogeneity. The forest plot revealed a correlation of the rs2073618 SNP with OTSC, yielded a significant summary (OR of 0.826, 95% CI [0.691–0.987], p = 0.035, Fig. 3). In addition, the combined results suggested a robust and significant association of the rs2073618 (c.9C > G) with OTSC under different genetic models: dominant model p = 0.006, OR = 0.701, 95% CI [0.545–0.901], recessive model p = 0.076, OR = 0.737, 95% CI [0.527–1.032], heterozygous model p = 0.022, OR = 0.729, 95% CI (0.557–0.955), and homozygous model p = 0.02, OR = 0.647, 95% CI [0.449–0.933].
Finally, sensitivity analysis was performed to reveal the influence of each individual study on the overall meta-analysis correlation. No single study affected the combined OR significantly, suggesting that this meta-analysis is relatively stable. Begg’s funnel plot (Fig. 4) and Egger regression test (p = 0.63) showed no publication bias within the studies included in the meta-analysis.
Discussion
Metabolic bone diseases and injuries of bones are major causes of human skeletal malformations resulting in abnormal mineralized tissue microarchitecture. These are serious health concerns with a severe socio-economic impact [36,37,38]. Amongst others, OTSC affects several million individuals worldwide with a late-onset of HL and represents a major problem that deserves greater attention. In recent years, a lot of effort has been made to identify the disease-causing genes of OTSC, resulting in the determination of ten loci [39, 40]. Map** these monogenic loci has not resulted in the recognition of any OTSC causative gene to date. Only a genome-wide analysis study identified the genetic association of the RELN gene with OTSC [10].
OPG is a glycoprotein which inhibits osteoclast formation, maturation, osteolysis and induces the apoptosis of activated cells. The main function of OPG is to regulate the normal bone turnover with a balanced bone resorption and formation. OPG is secreted by osteoblasts and mesenchymal stem/stromal cells in order to protect the skeleton from excessive bone resorption by binding to RANKL and preventing it from interacting with RANK, the osteoclastic cell surface receptor [45], showing the presence of the TGFβ superfamily in active otosclerotic foci. In addition, variants in the RELN gene appear to be associated with OTSC in a number of studies [8, 26, 46, 47] including British, Italian, Belgian-Dutch and Tunisian populations. Within this study, a multilocus association of six SNPs, including rs2073618 in OPG, rs1800472 in TGFβ1, rs39335, rs39350 and rs39374 in RELN and rs494252 in chromosome 11, revealed significant allele combinations with sex-specific association in OTSC subjects. A previous study assessed the relationship between OPG variants and bone mineral density (BMD) or osteoporotic fractures in postmenopausal Chinese. The study showed a significant association between rs2073618 and both BMD and osteoporotic fractures [48]. In addition, the same SNP in the OPG gene appears to be associated with a decreased BMD in a case-control study performed in Mexican-Mestizo women with rheumatoid arthritis [49]. Taken together, alteration in the OPG gene is related to abnormal bone metabolism and a number of skeletal pathologies including not only OTSC but also age-related bone diseases such as osteoporosis and rheumatoid arthritis.
To the best of our knowledge, this is the first study that shows an association between a SNP in the OPG gene and OTSC in a North African population, and supports previous reports of OTSC in an Indian population. Further investigations include functional validation of the rs2073618 SNP through in vivo insertion of the OPG promoter and monitor whether it affects DNA-protein complex formation and promoter activity. In addition, with regard to the importance and synergy of the RANKL/RANK/OPG in bone turnover, the remaining genes should be investigated in OTSC case-control association studies.
Conclusions
In conclusion, we evaluated the association between the OPG SNPs and OTSC and found that the rs3102734 and rs2073618 SNPs are linked to the onset of OTSC. This further supports the hypothesis that OPG may play an important role in bone turnover and metabolism within the otic capsule, and those functional polymorphisms within this gene may lead to the development of bone malformations such as OTSC.
Availability of data and materials
The data that support the findings of this study are not publicly available. Data are however available from the corresponding author upon reasonable request.
Abbreviations
- ACE:
-
Angiotensin-converting enzyme
- AGT:
-
Angiotensinogen
- BMD:
-
Bone mineral density
- BMP2:
-
Bone morphogenetic protein 2
- BMP4:
-
Bone morphogenetic protein 4
- CI:
-
Confidence interval
- COL1A1:
-
Collagen type I, alpha 1
- EDTA :
-
Ethylenediaminetetraacetic acid
- EM:
-
Expectation–maximization
- FGF2:
-
Fibroblast growth factor 2
- HL:
-
Hearing loss
- HWE:
-
Hardy-Weinberg equilibrium
- LD:
-
Linkage disequilibrium
- MEPE :
-
Matrix extracellular phosphoglycoprotein
- OPG:
-
Osteoprotegerin
- OR:
-
Odds ratio
- OTSC:
-
Otosclerosis
- PCR-RFLP:
-
Polymerase chain reaction-restriction fragment length polymorphism
- RANK:
-
Receptor activator of nuclear kappa-B
- RANKL:
-
Receptor activator of nuclear kappa-B ligand
- RELN:
-
Reelin
- SERPINF1:
-
Serpin family F member 1
- SNP:
-
Single nucleotide polymorphism
- TGFβ1:
-
Transforming growth factor beta 1
- TNF:
-
Tumor necrosis factor
- TRAIL:
-
Tumor necrosis factor related apoptosis inducing ligand
References
Ealy M, Smith RJH. The genetics of otosclerosis. Hear Res. 2010;266:70–4.
Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–55.
Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–49.
Rodan GA. Introduction to bone biology. Bone. 1992;13(Suppl 1):S3–6.
Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–45.
Fowler EP. Otosclerosis in identical twins. A study of 40 pairs. Arch Otolaryngol. 1966;83:324–8.
Morrison AW. Genetic factors in otosclerosis. Ann R Coll Surg Engl. 1967;41:202–37.
Mowat AJ, Crompton M, Ziff JL, Aldren CP, Lavy JA, Saeed SR, et al. Evidence of distinct RELN and TGFB1 genetic associations in familial and non-familial otosclerosis in a British population. Hum Genet. 2018;137:357–63.
Schrauwen I, Van Camp G. The etiology of otosclerosis: a combination of genes and environment. Laryngoscope. 2010;120:1195–202.
Schrauwen I, Ealy M, Huentelman MJ, Thys M, Homer N, Vanderstraeten K, et al. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. Am J Hum Genet. 2009;84:328–38.
Ziff JL, Crompton M, Powell HR, Lavy JA, Aldren CP, Steel KP, et al. Mutations and altered expression of SERPINF1 in patients with familial otosclerosis. Hum Mol Genet. 2016;25:2393–403.
Schrauwen I, Valgaeren H, Tomas-Roca L, Sommen M, Altunoglu U, Wesdorp M, et al. Variants affecting diverse domains of MEPE are associated with two distinct bone disorders, a craniofacial bone defect and otosclerosis. Genet Med. 2019;21:1199–208.
Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–19.
Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–7.
Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci U S A. 1998;95:3597–602.
Tsuda E, Goto M, Mochizuki S, Yano K, Kobayashi F, Morinaga T, et al. Isolation of a novel cytokine from human fibroblasts that specifically inhibits osteoclastogenesis. Biochem Biophys Res Commun. 1997;234:137–42.
Yasuda H, Shima N, Nakagawa N, Mochizuki SI, Yano K, Fujise N, et al. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology. 1998;139:1329–37.
Kang JH, Ko HM, Moon JS, Yoo HI, Jung JY, Kim MS, et al. Osteoprotegerin expressed by osteoclasts: an autoregulator of osteoclastogenesis. J Dent Res. 2014;93:1116–23.
Ney JT, Fehm T, Juhasz-Boess I, Solomayer EF. RANK, RANKL and OPG expression in breast Cancer – influence on osseous metastasis. Geburtshilfe Frauenheilkd. 2012;72:385–91.
Boyce BF, **ng L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys. 2008;473:139–46.
Zehnder AF, Kristiansen AG, Adams JC, Kujawa SG, Merchant SN, McKenna MJ. Osteoprotegrin knockout mice demonstrate abnormal remodeling of the otic capsule and progressive hearing loss. Laryngoscope. 2006;116:201–6.
Zehnder AF, Kristiansen AG, Adams JC, Merchant SN, McKenna MJ. Osteoprotegerin in the inner ear may inhibit bone remodeling in the otic capsule. Laryngoscope. 2005;115:172–7.
Iossa S, Morello G, Esposito T, Corvino V, Giannini P, Salvato R, et al. Exclusion of TNFRSF11B as candidate gene for Otosclerosis in Campania population. Indian J Otolaryngol Head Neck Surg. 2014;66:297–301.
Priyadarshi S, Ray CS, Biswal NC, Nayak SR, Panda KC, Desai A, et al. Genetic association and altered gene expression of osteoprotegerin in otosclerosis patients. Ann Hum Genet. 2015;79:225–37.
Khalfallah A, Schrauwen I, Mnejja M, HadjKacem H, Dhouib L, Mosrati MA, et al. Association of COL1A1 and TGFB1 polymorphisms with otosclerosis in a Tunisian population. Ann Hum Genet. 2011;75:598–604.
Khalfallah A, Schrauwen I, Mnaja M, Fransen E, Lahmar I, Ealy M, et al. Genetic variants in RELN are associated with otosclerosis in a non-European population from Tunisia. Ann Hum Genet. 2010;74:399–405.
Ugozzoli L, Wallace RB. Allele-specific polymerase chain reaction. Methods. 1991;2:42–8.
Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–14.
Armitage P, Berry G, Matthews JNS. Statistical methods in medical research: Wiley; 2001. https://www.wiley.com/en-au/Statistical+Methods+in+Medical+Research%2C+4th+Edition-p-9780632052578.
Borenstein M, Hedges LV, Higgins JP, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. 2010;1:97–111.
Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206.
** C, Hu Y, Chen XC, Zheng FY, Lin F, Zhou K, et al. Laparoscopic versus open myomectomy--a meta-analysis of randomized controlled trials. Eur J Obstet Gynecol Reprod Biol. 2009;145:14–21.
Bedi U, Singh M, Singh P, Molnar J, Khosla S, Arora R. Effects of statins on progression of coronary artery disease as measured by intravascular ultrasound. J Clin Hypertens (Greenwich). 2011;13:492–6.
Jiang K, Lepak DP, Hu J, Baer JC. How does human resource management influence organizational outcomes? A meta-analytic investigation of mediating mechanisms. Acad Manag J. 2012;55:1264–94.
Vyas A, Swaminathan PD, Zimmerman MB, Olshansky B. Are treatments for vasovagal syncope effective? A meta-analysis. Int J Cardiol. 2013;167:1906–11.
Woolf AD, Akesson K. Understanding the burden of musculoskeletal conditions. The burden is huge and not reflected in national health priorities. BMJ. 2001;322:1079–80.
DALYs GBD, Collaborators H. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2017;390:1260–344.
Briggs AM, Woolf AD, Dreinhofer K, Homb N, Hoy DG, Kopansky-Giles D, et al. Reducing the global burden of musculoskeletal conditions. Bull World Health Organ. 2018;96:366–8.
Weegerink NJ, Schrauwen I, Huygen PL, Pennings RJ, Cremers CW, Van Camp G, et al. Phenotype of the first otosclerosis family linked to OTSC10. Laryngoscope. 2011;121:838–45.
Ealy M, Smith RJH. Otosclerosis. Adv Otorhinolaryngol. 2011;70:122–9.
Boyce BF, **ng L. The RANKL/RANK/OPG pathway. Curr Osteoporos Rep. 2007;5:98–104.
Karosi T, Csomor P, Szalmas A, Konya J, Petko M, Sziklai I. Osteoprotegerin expression and sensitivity in otosclerosis with different histological activity. Eur Arch Otorhinolaryngol. 2011;268:357–65.
Lehnerdt G, Unkel C, Metz KA, Jahnke K, Neumann A. Immunohistochemical evidence of BMP-2, −4 and −7 activity in otospongiosis. Acta Otolaryngol. 2008;128:13–7.
Frenz DA, Liu W, Capparelli M. Role of BMP-2a in otic capsule chondrogenesis. Ann N Y Acad Sci. 1996;785:256–8.
Liu W, Oh SH, Kang Yk Y, Li G, Doan TM, Little M, et al. Bone morphogenetic protein 4 (BMP4): a regulator of capsule chondrogenesis in the develo** mouse inner ear. Dev Dyn. 2003;226:427–38.
Sommen M, Van Camp G, Liktor B, Csomor P, Fransen E, Sziklai I, et al. Genetic association analysis in a clinically and histologically confirmed otosclerosis population confirms association with the TGFB1 gene but suggests an association of the RELN gene with a clinically indistinguishable otosclerosis-like phenotype. Otol Neurotol. 2014;35:1058–64.
Iossa S, Corvino V, Giannini P, Salvato R, Cavaliere M, Panetti M, et al. The rs39335 polymorphism of the RELN gene is not associated with otosclerosis in a southern Italian population. Acta Otorhinolaryngol Ital. 2013;33:320–3.
Sheng X, Cai G, Gong X, Yao Z, Zhu Y. Common variants in OPG confer risk to bone mineral density variation and osteoporosis fractures. Sci Rep. 2017;7:1739.
Nava-Valdivia CA, Saldana-Cruz AM, Corona-Sanchez EG, Murillo-Vazquez JD, Moran-Moguel MC, Salazar-Paramo M, et al. Polymorphism rs2073618 of the TNFRSF11B (OPG) gene and Bone mineral density in Mexican women with rheumatoid arthritis. J Immunol Res. 2017;2017:7680434.
Acknowledgments
We gratefully thank all the subjects for their collaboration in achieving this study.
Funding
This work was supported by a research grant under the Tunisia/India agreement on science and technology cooperation [EPIGENOTOS]. The funding source did not play any roles in the study design, data collection, analysis and interpretation, and writing the manuscript.
Author information
Authors and Affiliations
Contributions
AB, AT: Conceived the study, performed the experiments, analyzed and interpreted the data, and wrote the manuscript draft. FJ: Assisted and performed additional experiments. AC, IA, AG, IC: ascertained patients, diagnosis and sample collection. KH, NS: Contributed in the data analysis. AS, MAM: Contributed in the samples’ data collection. PVR: Assisted in the analysis and interpretation of the data, and revised the manuscript. SM: Designed the work, contributed in the data analysis, revised the manuscript, and acquired the funding of the project. All the authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethical committee of the University Hospital of Sfax, and written informed consent was obtained from all participants of this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bouzid, A., Tekari, A., Jbeli, F. et al. Osteoprotegerin gene polymorphisms and otosclerosis: an additional genetic association study, multilocus interaction and meta-analysis. BMC Med Genet 21, 122 (2020). https://doi.org/10.1186/s12881-020-01036-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-020-01036-8