Abstract
Background
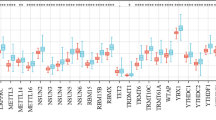
N7-methylguanosine (m7G) is present in a wide variety of organisms and has important roles. m7G has been reported to be involved in multiple biological processes, and recent studies have reported that changes in RNA modifications result in tumor cellular transformation and cancer, such as colon adenocarcinoma, lung cancer, and intrahepatic cholangiocarcinoma. However, little is known about the function of the m7G in colon adenocarcinoma.
Methods
We established two clusters based on the expression of all genes associated with m7G to explore the expression pattern of 31 key regulatory factors of m7G RNA and assess the prognostic value of regulatory factors. Wilcoxon test and differential box line plots were applied for bioinformatics analysis. Receiver Operating and Kaplan‒Meier curves were utilized to evaluate the prognostic value. Finally, four genes' expression in the colon cancer cell line was confirmed by qRT-PCR.
Results
From The Cancer Genome Atlas database, we found that the expression levels of 25 out of the 31 key N7-methylguanosine RNA modification regulators were significantly different in colon adenocarcinoma. According to 25 methylation regulators’ expression, we identified two subgroups by consensus clustering, in which the prognosis was worse in Group 2 than in Group 1 and was significantly correlated with age. Cluster 2 was significantly enriched in tumor-associated pathways, and immune cells were highly infiltrated in Cluster 1 but weakly infiltrated in Cluster 2. Further results indicated that this risk profile may serve as a standalone predictive factor for colon adenocarcinoma, and the four genetic risk profiles’ prognostic relatedness was successfully verified through Gene Expression Omnibus dataset. At last, A nomogram for prognosis was created according to age, sex, histological grading, clinicopathological staging, and hazard score to accurately predict patient prognosis in colon adenocarcinoma. We successfully validated the differential expression of four genes using qRT-PCR.
Conclusions
In the present study, we revealed the important contribution of key regulators associated with m7G RNA modifications based on all gene expression in colon adenocarcinoma and developed a signature of risk that serves as a promising prognostic marker for patients with colon adenocarcinoma.
Similar content being viewed by others
Introduction
Global Cancer Statistics 2020 reported that the incidence of colorectal cancer was 10% and that the mortality rate was 9.4%, among all cancers, it is the second most common cause of mortality [1]. Up to 20–30% of colon cancer patients with early-stage illness will develop distant metastases despite complete segmental resection [2]. Colon cancers are not only anatomically different from rectal cancers but also pathologically require different staging procedures. Furthermore, colon and rectal cancers require different neoadjuvant treatments and compatible surgical approaches [3]. The current treatment modalities for colon cancer mainly include surgery and radiotherapy. Although surgery is usually sufficient for early-stage colon cancer, advanced colon cancer requires a combination of preoperative and postoperative radiotherapy [4]. Despite improvements in systemic therapy, the five-year survival rate for CRC metastatic disease patients is around 12–14% [5]. Moreover, patients who are not diagnosed promptly at an early stage often end up with poor treatment outcomes and a poor prognosis. With the development of RNA epigenetics, it has become easier to identify novel biomarkers and therapeutic targets, Moreover, mechanisms of RNA epigenetics may be crucial for improving early cancer diagnosis, treatment, and prognosis.
A regular RNA methylation alteration called m7G has a significant impact on the entire biological process. RNA methylation is a basic process of epigenetic regulation. A large amount of evidence shows that the methylation of RNA plays a crucial function in many biological processes, and RNA methylation’s dysregulation is highly correlated with the development of human cancers, especially gastrointestinal tumors [3A–C). We used principle component analysis (PCA) to further investigate the two classes for the purpose of verifying clustering results. Significant differences between Clusters 1 and 2 were visible in PCA plots (Fig. 3D). We next evaluated whether there were notable variations in clinical parameters and overall survival (OS) between these two groups. As a consequence, OS in cluster 1 was considerably superior to OS in cluster 2 (p < 0.01) (Fig. 4B). Furthermore, Cluster 2 showed upregulated expression levels of most RNA modification regulator genes rather than Cluster 1 (Fig. 4A). Although there were no significant variations in histological grading, pathological stage, or sex, age variances between the two grou**s were noticeably different. (p < 0.05) (Fig. 4A). Consensus clustering's findings, therefore, showed a strong correlation between colon adenocarcinoma's malignancy and patterns of expression of RNA modification regulators in m7G.
Consensus cluster analysis of COAD. A Subgroup correlation when using the k = 2 clustering factor. B For k = 2–9, the cumulative distribution function (CDF) is shown in the picture. C In the CDF curve for k = 2–9, the corresponding variance is in the area below the CDF curve. D An analysis of RNA-seq data based on principal components. Blue dots indicate low-risk clusters, whereas red dots indicate high-risk clusters
Differences in clinical and pathological characteristics of Clusters 1 and 2 as well as overall survival. A Here is a heatmap showing the clinicopathological characteristics of these two clusters. Red and green represent high and low expression, respectively. The differences in age were significant (p < 0.05). B Among Clusters 1 and 2, an OS Comparison is made. *p < 0.05; **p < 0.01; ***p < 0.001. OS, overall survival
We next performed GO and KEGG analyses of the differentially expressed genes between Clusters 1 and 2 to further explain the outcomes of clustering in terms of underlying biological processes. According to analysis of GO, genes which are downregulated were mainly involved in biological processes connected to cancer listed below: neutrophil chemotaxis and migration; antimicrobial humoral immune response mediated by an antimicrobial peptide; antimicrobial humoral response; response to chemokine; cellular response to chemokine and humoral immune response; and response to chemokine (Fig. 5A, B). In addition, according to KEGG analysis, the upregulated genes are associated with cytokine-cytokine receptor interactions, and the majority of the downregulated genes were associated with COVID-19, a coronavirus illness (Fig. 5C, D).
Development of a prognostic risk model for m7G RNA modification-related genes based on the expression levels of all genes
Our study performed univariate Cox regression in order to determine whether expression levels of the important regulators are connected to a patient's prognosis for colon cancer. The findings demostrated that OS as well as 16 of these genes existed significant conection (p < 0.01) (Fig. 6A). Among these 16 regulators, GABBR1, LINC00174, HSF4, LTB4R, EXOC3L4, RPL32P3, MAN2C1, YJEFN3, ZNF692, UPK3B, DNAH1, ZNF767P, MTMR9LP, AGAP9, L3HYPDH, and ADAMTS13 were considered risk genes with HR (hazard ratio) > 1. Among them, LINC00174, MAN2C1, DNAH1, MTMR9LP, AGAP9, and ADAMTS13 were at higher risk with HR > 2. Subsequently, all gene-modifying regulators with the highest prognostic ability were screened by LASSO Cox regression analysis (Fig. 6B, C), which identified the following four genes to estimate the risk of colon adenocarcinoma: heat shock transcription factor 4 (HSF4); uroplakin 3B (UPK3B); zinc finger family member 767, pseudogene (ZNF767P); and ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 (AGAP9) (Fig. 6D). Calculation of risk score in following formula: risk score = (0.0859 HSF4 expression value) + (0.3854 UPK3B expression value) + (0.0592 ZNF767P expression value) + (0.1646 AGAP9 expression value). The four genes highly expressed in tumor cells were verified by qRT-PCR (Fig. 6E). Additionally, Fig. 6F shows the risk distribution score of patients with colon adenocarcinomas, and each patient's survival status was shown using a scatter plot (dot plot) (Fig. 6G).
Establishment of a model for predictive disease risk based on the regulator genes of RNA modification genes in m7G. A Analysis of the regulator genes correlated with m7G RNA methylation using univariate Cox regression. B–D The procedure utilized to create the signature with Cox regression and the absolute shrinkage and selection operator (LASSO). E Expression levels of the four genes in FHC (normal) and SW480 (tumor) cells. F Risk score distributions of the risk score model. G A prognostic model of survival status distributions. *p < 0.05; **p < 0.01; ***p < 0.001
In line with median risk scores, colon cancer patients were divided into low and high risk groups to investigate the four-gene signature model's prognostic effects. According to the survival analysis, there was a worse overall survival rate (OS) for patients with high-risk scores than for those with low-risk scores (Fig. 7A, p < 0.001). In the high-risk group, the five-year OS rate was 56.3%, while in the low-risk cluster, the five-year OS rate was 73.3%. The area under the curve (AUC) values for the one-year, two-year, three-year, and five-year OS were 0.648, 0.663, 0.670, and 0.628, respectively, as determined by ROC curve analysis, which indicated the strong predictive potential for survival outcomes (Fig. 7B).
Prognostic model survival analyses based on Kaplan–Meier curves. Using the median risk score as the cutoff, patients from the two datasets were divided into low-risk (blue) and high-risk (yellow) groups. A In TCGA cohort, the group which is in low risk had a higher probability of survival compared to the group which is in high risk (p < 0.001). B The one-, two-, three- and five-year AUC values were 0.648, 0.663, 0.670, and 0.628, in the TCGA cohort. C In GEO cohort, the model for prognosis was verified to be accurate. The low-risk group's chances of survival were higher than those of the high-risk group (p = 0.04). D with one-, two-, three-, and five-year AUC values of 0.557, 0.617, 0.529, and 0.535, respectively. GEO Gene Expression Omnibus, TCGA The Cancer Genome Atlas, AUC area under the curve
Using GEO's database to validate predictive signatures
GEO's database microarray data (GSE39582) as a testing set were conducted to evaluate the four-gene signature's prognostic value. According to the cut-off values of the TCGA cohort, 228 individuals with colon adenocarcinoma in the GSE39582 cohort were grouped into two groups. 117 people were in the category of having high risk. About another 111 people were defined as low risk. Survival analysis showed that patients with colon cancer in the low-risk group had a considerably better OS than patients in the high-risk group, which is in line with the findings in the TCGA cohort. (Fig. 7C , p = 0.04). In the one-, two-, three-, and five-year OS, the AUC values were 0.557, 0.617, and 0.535., which demonstrated that the prediction model accurately predicted the OS of colon adenocarcinoma patients (Fig. 7D).
Prediction of colon cancer patient prognosis using the four-gene risk signature
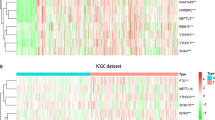
Comparing low and high risk groups based on pathological stage N revealed significant differences in clinical parameters (p < 0.001). The heatmap shows the expression and clinical correlation of four genes, and all four genes are strongly associated with prognosis between the two groups. (Fig. 8A). In total, 458 cases were added to the Cox regression analysis after cases with insufficient clinical information were eliminated. According to univariate analysis, OS is significantly associated with four-gene risk scores, T stage, N stage, and clinicopathological stage in patients with colon adenocarcinoma (Fig. 8B , p < 0.001). In order to ascertain if four-gene risk marker is a predictive marker for colon adenocarcinoma independent of the other clinicopathological characteristics, multivariate Cox regression analysis was performed. The results revealed that OS in individuals with colon adenocarcinoma was independently correlated with risk score and clinicopathological stage. (Fig. 8C , p < 0.001). These findings indicated that the four-gene risk signature can be utilized as an indicator which can independently prognosticate for colon adenocarcinoma regardless of sex, age, histological grade, and pathological stage.
The prediction ability of the risk score and clinicopathological characteristics for COAD patient prognosis. A The heatmap displays the expression of five m7G RNA modification regulators and the distribution of clinicopathological characteristics in high- and low-risk groups. In the heatmap, it displays the expression levels of five m7G RNA modification regulators, as well as the clinicopathological characteristics’ distribution in groups at high- and low-risk. B Clinicopathological parameter and OS assessments using univariate Cox regression. C Clinicopathological variables and OS were analyzed using multivariate Cox regression. *p < 0.05; **p < 0.01; ***p < 0.001. COAD colon adenocarcinoma, OS overall survival
Differential analysis of immune cell infiltration and immune function in groups at high- and low-risk
We examined the variations in immune cell infiltration connected to colon cancer in the TCGA database. Macrophage, neutrophil, and regulatory T cells (Tregs) infiltration levels were high in group which is at low risk but low in the group which is at high risk (p < 0.001) (Fig. 9A). Differences in immune functions associated with colon adenocarcinoma were analyzed. The levels of APC coinhibition, APC costimulation, chemokine receptors (CCRs), cytolytic activity, inflammation promotion, parainflammation, and type-II IFN-response were high in the low-risk group but low in the high-risk group (p < 0.001) (Fig. 9B).
Analysis of immunological differences between high- and low-risk groups of COAD patients. Groups at low risk are represented by the blue box, while groups at high risk are represented by the red box. The horizontal line in the box showed the median value, which is the expression of different groups. A The levels of macrophages, neutrophils, and regulatory T cells (Tregs) were high in the group at low risk but low in the group at high risk (p < 0.001). B In contrast to the group at high risk, the levels of APC coinhibition, APC costimulation, and CCR were low in the group at low risk (p < 0.001). C The levels of macrophages, neutrophils, and Tregs were significantly higher in the group at low risk and significantly lower in the group at high risk (p < 0.001). D The levels of CCRs, cytolytic activity, and inflammation promotion were high in the group at low risk but low in the group at high risk. *p < 0.05; **p < 0.01; ***p < 0.001. COAD, colon adenocarcinoma; CCRs, chemokine receptors
Immune cell differential analysis was also verified to be significant in the GEO database with high infiltration levels of macrophages, neutrophils, and Tregs in the group which is at low risk but low infiltration of these cells in the group which is at high risk(p < 0.001) (Fig. 9C). Regarding immune function differential analysis, the levels of CCRs, cytolytic activity, and inflammation promotion were high in the group at low risk but low in the group at high risk (p < 0.001) (Fig. 9D).
Construction of a nomogram for colon adenocarcinoma prognosis
To establish a quantitative method to predict individual survival, we created a novel predictive nomogram based upon age, sex, histological grade, pathological stage, and risk score. (Fig. 10). The outcomes demonstrated that in patients with colon adenocarcinoma, one, three, and five-year OS were systematically predicted by the nomogram.
Discussion
Based on four modifiers of key regulators closely correlated with prognosis in patients with colon adenocarcinoma, a predictive model was constructed by us. Patients with colon adenocarcinoma can have their prognosis reliably predicted using the established risk score. After selecting patient data from TCGA database that satisfied the inclusion requirements, we first performed a bioinformatics analysis. To find genes that were differently expressed, we performed the Wilcoxon test in R. And these genes can encode regulators of m7G RNA methylation. In colon adenocarcinoma, we then identified the differentially expressed m7G RNA modification regulators. Using the screened gene expression patterns in accordance with k = 2, we divided the colon cancer patients into two groups with distinct clinical outcomes. We also analyzed the variations in immune function and immune cell infiltration between the two groups. Following these procedures, we created the predictive risk model which is based upon all levels of gene expression. Finally, to validate the prognostic model, we repeated the above steps to validate the prognostic model by screening the data of colon adenocarcinoma patients who met the inclusion criteria through the GEO database, which was well validated. Based on the accuracy of the results, we evaluated the prognosis of colon adenocarcinoma patients by generating a nomogram.
Heat shock transcription factor 4 (HSF4) is a heat shock factor and is a member of the HSF family. HSF4 has various physiological functions as follows: regulating the transcriptional program of the heat response or stress response; regulating cell proliferation and differentiation during development; regulating DNA damage repair, and regulating normal physiological processes. HSF4’s Alterations are also strongly linked to cataracts, cancer, and other illnesses [21]. Mice lacking HSF4 produce irregular lenses and develop cataracts early in experiments [22,23,24]. A previous study on a cohort of patients with congenital cataracts in China has indicated that disease development is closely associated with genetic mutations in HSF4 DBD [25]. By playing a critical role in carcinogenesis and tumor progression, HSF4 has been demonstrated to enhance EMT by activating the AKT pathway in a HIF1α-dependent manner in hepatocellular carcinoma; Hepatocellular carcinoma cells are better able to migrate, disseminate, and invade when HSF4 is upregulated, which promotes aggressive tumor behavior, indicating that high HSF4 expression may be a predictor of poor hepatocellular carcinoma after radical resection [26].
The expression of uroplakin 3B (UPK3B), Several tissues, and organs have been shown to include a few of the main structural elements of uroepithelial tissue (UPK3A and UPK3B). For example, in mouse embryos, Cre recombinase activity driven by UPK3B is detected in the liver, heart, kidney, lung, and neural crest cells. UPK3B expression has been detected in mouse testes, epididymal spermatozoa, ovarian follicles, and oviductal mucosa, proving that UPK3B may be extremely important for the development of mouse gametes as well as gamete delivery organs [27]. A whole transcriptome analysis of placental changes in fetuses with prenatal arsenic exposure has reported that UPK3B is one of the most significant ‘off’ genes for arsenic exposure in females [28]. Transcriptome analysis of liver fibrosis has identified UPK3B as a potential regulator of hepatic stellate cell (HSC) activation-induced liver fibrosis [29]. Low FOXA1 expression has been linked to earlier tumor staging, while FOXA1 deletion has been associated with high histological grade. Increased UPK3B expression, decreased E-calcineurin expression, and increased cell proliferation have been observed in FOXA1-deficient RT4 bladder cancer cells, demonstrating a strong relationship between high UPK3B expression and tumor malignancy [30].
These recent studies have demonstrated that HSF4 and UPK3B are both closely associated with tumors. Although few studies have been reported on the ZNF767P and AGAP9 genes, they are promising research targets. What’s more, by analyzing immune cell infiltration and immune activity differently, macrophages contribute a significant part in the formation of tumors too. while the therapeutic effect can also be achieved through the modulatory role of engineered macrophages in the tumor immune microenvironment and tumor therapy [31]. A dysregulation of macrophage-mediated immunosuppression leads to chronic inflammation at low grade due to tissue-specific macrophages and neutrophils, which ultimately leads to the development of cancer [32]. Neutrophils are considered complex cells with many specific functions, and they act as effectors of the innate immune response and play a regulatory role in multiple processes, such as cancer, acute injury, repair, autoimmunity, and chronic inflammation [33]. In the host, neutrophils reflect inflammation, which is a hallmark of cancer [34]. An association between high layilin (LAYN) expression and poor overall survival in colon cancer patients has been demonstrated by Pan et al.. A positive correlation exists between LAYN expression and macrophage and neutrophil infiltration in colon adenocarcinoma (COAD) [35]. The immune system tightly controls Th17/Treg homeostasis through the TGF-/IL-2 and IL-6 cytokine axis. Regulatory T cells (Tregs) are necessary for self-tolerance and defense against autoimmunity, and they are typically linked to the advancement of cancer [36]. By maintaining Treg activity and accumulation in the colon, glycoprotein-A repetition predominant (GARP) reduces cancer immunity [37]. Thus, these findings indicate that macrophages, neutrophils, and Tregs are highly infiltrated in the group at low risk, which reduces the tumorigenesis development.
According to the current study, three immune function differential analyses were validated in both TCGA and GEO databases. Numerous elements of cancer biology have been identified to involve chemokines and their receptors; their possible targets have been evaluated in several studies, and chemokine receptor inhibitors have been used in clinical practice in hematologic malignancies [38]. In patients with gastric cancer, the cytolytic activity score can be employed as a biomarker in antitumor immunity and clinical prognosis [39] but also to evaluate anticancer immunity in colorectal cancer [40]. Nuclear factor-κB (NF-κB), which promotes inflammation, is a central mediator of the inflammatory process, and activation of NF-κB is also prevalent in cancer, which is mainly driven by inflammatory cytokines in the tumor microenvironment [41]; thus, inflammation promotion plays a crucial role in tumorigenesis. In conclusion, chemokine receptors, cytolytic activity, and inflammation promotion are closely related to tumors and play critical roles in the diagnosis, treatment, and prognosis of tumors.
The present study had several benefits. First, based on the patterns of major regulators related to m7G RNA alterations expressed in all genes, we generated the first predictive model for colon cancer. Second, the model was constructed using a variety of statistical techniques, and both the test cohort and the entire cohort were used for validation. As a result, the predictive risk model for patients with colon cancer is precise and trustworthy. The accuracy of the risk score model in predicting OS was higher than that of pathological stage and age, and the risk score model can be employed as a standalone prognostic indicator. Finally, throughout the advancement of colon cancer research, our model can also be utilized to predict immune cell infiltration and to study differential immune function. However, the present study had several limitations. First, we generated an unvalidated prognostic risk model based on a public database rather than a clinical study. In addition, the possible mechanisms through which the important regulators of m7G RNA modification affect colon cancer progression need to be further investigated by basic experiments. Further, there was a lack of in vivo or in vitro experiments exploring the molecular functions of the four genes in the model. Further studies are required to elucidate the mechanism.
Conclusions
In the present study, a four-gene signature of colon cancer, consisting of HSF4, UPK3B, ZNF767P, and AGAP9, was generated and validated. It can be used as an auxiliary predictive variable, and patients with colon adenocarcinoma can be predicted how long their survival will last using this analysis.
Availability of data and materials
All datasets presented in this study are included in the article/supplementary material. All the R packages above are available from http: //www.Bioconductor.org.
Abbreviations
- m7G:
-
N7-methylguanosine
- COAD:
-
Colon adenocarcinoma
- ROC:
-
Receiver operating characteristic curves
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- FC:
-
Fold change rate
- FDR:
-
False discovery rate
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene ontology
- LASSO:
-
Selection operator Cox regression
- GSEA:
-
Gene set enrichment analysis
- PCA:
-
Principal component analysis
- OS:
-
Overall survival
- CDF:
-
The cumulative distribution function
- HR:
-
Hazard ratio
- AUC:
-
Area under curve
References
Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008;2008(3):cd005390.
Tamas K, et al. Rectal and colon cancer: not just a different anatomic site. Cancer Treat Rev. 2015;41(8):671–9.
Chakrabarti S, et al. Early stage colon cancer: current treatment standards, evolving paradigms, and future directions. World J Gastrointest Oncol. 2020;12(8):808–32.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
**e S, et al. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int. 2020;20(1):585.
Wang H, et al. A brief review of machine learning methods for RNA methylation sites prediction. Methods. 2022;203:399–421.
Chen X, et al. RNA methylation and diseases: experimental results, databases, Web servers and computational models. Brief Bioinform. 2019;20(3):896–917.
Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. 2014;31:1–7.
Liu N, Pan T. RNA epigenetics. Transl Res. 2015;165(1):28–35.
Teng PC, et al. RNA modifications and epigenetics in modulation of lung cancer and pulmonary diseases. Int J Mol Sci. 2021;22(19):10592.
Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8(10):1253–66.
Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11(5):821–30.
Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2(5):611–31.
Pandolfini L, et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol Cell. 2019;74(6):1278-1290.e9.
Orellana EA, et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81(16):3323-3338.e14.
Liu Y, et al. Overexpressed methyltransferase-like 1 (METTL1) increased chemosensitivity of colon cancer cells to cisplatin by regulating miR-149-3p/S100A4/p53 axis. Aging (Albany NY). 2019;11(24):12328–44.
Dai Z, et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81(16):3339-3355.e8.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Zhang L, et al. Expression pattern and prognostic value of key regulators for m6A RNA modification in hepatocellular carcinoma. Front Med (Lausanne). 2020;7:556.
Syafruddin SE, et al. More than meets the eye: revisiting the roles of heat shock factor 4 in health and diseases. Biomolecules. 2021;11(4):523.
Fujimoto M, et al. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23(21):4297–306.
Shi X, et al. Removal of Hsf4 leads to cataract development in mice through down-regulation of gamma S-crystallin and Bfsp expression. BMC Mol Biol. 2009;10:10.
Min JN, et al. Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis. 2004;40(4):205–17.
Bu L, et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31(3):276–8.
Ma P, et al. HSP4 triggers epithelial-mesenchymal transition and promotes motility capacities of hepatocellular carcinoma cells via activating AKT. Liver Int. 2020;40(5):1211–23.
Kuriyama S, Tamiya Y, Tanaka M. Spatiotemporal expression of UPK3B and its promoter activity during embryogenesis and spermatogenesis. Histochem Cell Biol. 2017;147(1):17–26.
Winterbottom EF, et al. Transcriptome-wide analysis of changes in the fetal placenta associated with prenatal arsenic exposure in the New Hampshire Birth Cohort Study. Environ Health. 2019;18(1):100.
Huang HM, et al. Histone deacetylase inhibitor givinostat alleviates liver fibrosis by regulating hepatic stellate cell activation. Mol Med Rep. 2021;23(5):305.
DeGraff DJ, et al. Loss of the urothelial differentiation marker FOXA1 is associated with high grade, late stage bladder cancer and increased tumor proliferation. PLoS ONE. 2012;7(5):e36669.
**a Y, et al. Engineering macrophages for cancer immunotherapy and drug delivery. Adv Mater. 2020;32(40):e2002054.
Jackaman C, et al. Aging and cancer: the role of macrophages and neutrophils. Ageing Res Rev. 2017;36:105–16.
Liew PX, Kubes P. The neutrophil’s role during health and disease. Physiol Rev. 2019;99(2):1223–48.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Pan JH, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6.
Knochelmann HM, et al. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol. 2018;15(5):458–69.
Salem M, et al. GARP dampens cancer immunity by sustaining function and accumulation of regulatory T cells in the colon. Cancer Res. 2019;79(6):1178–90.
Mollica Poeta V, et al. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Front Immunol. 2019;10:379.
Hu Q, et al. Cytolytic activity score as a biomarker for antitumor immunity and clinical outcome in patients with gastric cancer. Cancer Med. 2021;10(9):3129–38.
Narayanan S, et al. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25(8):2323–31.
DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246(1):379–400.
Acknowledgements
Ying Huang participated in the supplementary experimental design, suggested constructive modifications and participated in the revision of the article, and added important references.
Funding
Joint Funds for the innovation of Science and Technology, Fujian province (2017Y9038, 2019Y9101); Fujian Provincial Health Technology Project (2020GGB022); Natural Science Foundation of Fujian Province (2022J01753).
Author information
Authors and Affiliations
Contributions
YZ and ZZ wrote the main manuscript text. MT and JC prepared figures. SH and PC designed the study. YZ, ZZ, MT and JC performed the research. All authors reviewed the manuscript. YZ and ZZ contributed equally to the work. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The original code and visual data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhu, Y., Zhao, Z., Thandar, M. et al. Expression patterns and prognostic value of key regulators associated with m7G RNA modification based on all gene expression in colon adenocarcinoma. BMC Gastroenterol 23, 22 (2023). https://doi.org/10.1186/s12876-023-02657-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-023-02657-y