Abstract
Background
Myocardial ischemia-reperfusion injury (I/RI) is a major cause of perioperative cardiac-related adverse events and death. Studies have shown that sevoflurane postconditioning (SpostC), which attenuates I/R injury and exerts cardioprotective effects, regulates mitochondrial dynamic balance via HIF-1α, but the exact mechanism is unknown. This study investigates whether the PI3K/AKT pathway in SpostC regulates mitochondrial dynamic balance by mediating HIF-1α, thereby exerting myocardial protective effects.
Methods
The H9C2 cardiomyocytes were cultured to establish the hypoxia-reoxygenation (H/R) model and randomly divided into 4 groups: Control group, H/R group, sevoflurane postconditioning (H/R + SpostC) group and PI3K/AKT blocker (H/R + SpostC + LY) group. Cell survival rate was determined by CCK-8; Apoptosis rate was determined by flow cytometry; mitochondrial membrane potential was evaluated by Mito Tracker™ Red; mRNA expression levels of AKT, HIF-1α, Opa1and Drp1 were detected by quantitative real-time polymerase chain reaction (qRT-PCR); Western Blot assay was used to detect the protein expression levels of AKT, phosphorylated AKT (p-AKT), HIF-1α, Opa1 and Drp1.
Results
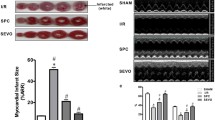
Compared with the H/R group, the survival rate of cardiomyocytes in the H/R + SpostC group increased, the apoptosis rate decreased and the mitochondrial membrane potential increased. qRT-PCR showed that the mRNA expression of HIF-1α and Opa1 were higher in the H/R + SpostC group compared with the H/R group, whereas the transcription level of Drp1 was lower in the H/R + SpostC group. In the H/R + SpostC + LY group, the mRNA expression of HIF-1α was lower than the H/R + SpostC group. There was no difference in the expression of Opa1 mRNA between the H/R group and the H/R + SpostC + LY group. WB assay results showed that compared with the H/R group, the protein expression levels of HIF-1α, Opa1, P-AKT were increased and Drp1 protein expression levels were decreased in the H/R + SpostC group. HIF-1α, P-AKT protein expression levels were decreased in the H/R + SpostC + LY group compared to the H/R + SpostC group.
Conclusion
SpostC mediates HIF-1α-regulated mitochondrial fission and fusion-related protein expression to maintain mitochondrial dynamic balance by activating the PI3K/AKT pathway and increasing AKT phosphorylation, thereby attenuating myocardial I/R injury.
Similar content being viewed by others
Introduction
Myocardial ischemia-reperfusion injury (I/RI) is a common clinical pathophysiological process that is the main reason for the occurrence and death of the perioperative adverse cardiac events and death [1]. Sevoflurane postconditioning (SPostC) has been confirmed to exert good myocardial protective effects against myocardial I/RI by inhibiting apoptosis, reducing myocardial LDH content, decreasing ROS production, and increasing ATP content, and is a potential treatment against myocardial injury in the perioperative period [2,12, 13]. It has been demonstrated that in the rat heart, SPostC exerts a cardioprotective effect against I/RI through activation of the PI3K/AKT signal pathway [14]. Numerous studies have shown that PI3K/Akt/HIF-1α pathway is frequently activated in human cancers and plays a key role in promoting glycolysis [15,Full size image
-
(1)
Control (Control) group: H9C2 cells incubated normally in the CO2 incubator for 48 h without any interventions.
-
(2)
Hypoxia/reoxygenation (H/R) group: H9C2 cells were incubated normally in CO2 incubator for 42 h and then placed in a tri-gas incubator for 3 h hypoxia and 3 h reoxygenation.
-
(3)
Sevoflurane postconditioning (H/R + SpostC) group: After 42 h of normal incubation in the CO2 incubator, H9C2 cells were placed in a tri-gas incubator for 3 h of hypoxia. Reoxygenation was started before SpostC for 15 min, followed by a further 165 min of incubation in the CO2 incubator.
-
(4)
PI3K/AKT inhibitor (H/R + SpostC + LY) group: After 40 h of normal incubation in the CO2 incubator, H9C2 cells were cultured in medium containing 10µM LY294002 until the end of the experiment, for 8 h. After 2 h of intervention, the cells were placed in a triple gas incubator for 3 h of hypoxia and SPostC for 15 min before the start of reoxygenation, followed by continued incubation in a CO2 incubator for 165 min.
Determination of cell viability
A single cell suspension of 5 × 104 cells/mL was prepared by digesting well-grown, 90% fused H9C2 cells with trypsin and cultured using complete medium. The suspension was inoculated into 96-well plates (100µL/well, 5 × 103Cells/well). Cells were incubated at 37 °C and 5% CO2 for 24 h and then grouped according to experiment. Discarded the medium, added 100µL of 10% CCK-8 solution to each well and continued to incubate in the incubator. After 1 h, measured the OD value at 450 nm with a microplate reader to detect the cell survival rate.
Flow cytometry assay
The percentage of cells apoptosis was determined by PE annexin-V apoptosis detection kit I. (BD Biosciences, USA) according to the manufacturer’s instructions as previously described (Ma et al., 2020).H9C2 cells in good growth condition with 90% confluence were digested with trypsin and prepared into a single cell suspension of 5 × 104 cells/mL in complete medium and inoculated into 6-well plates; after treatment in experimental groups, the culture fluid from each group of cell bottles was aspirated together with the cells into a centrifuge tube (containing apoptotic or necrotic cells already in suspension) and centrifuged at 1000 rpm for 5 min. The supernatant was discarded. Wash twice with pre-cooled PBS and discard the supernatant. Add 500µL of 1×Binding Buffer to resuspend the cells and pass through a 200 Mesh sieve to make a single cell suspension. Add 5µL of Annexin V-PE and 10µL of 7-AAD to each tube, mix gently and leave for 5 min at 4°C, protected from light. Flow cytometry was performed within 30 min.
Detection of mitochondrial membrane potential
We assessed mitochondrial membrane potential (Δψm) using Mito Tracker™ Red (Thermo Fisher Scientific, USA).H9C2 cells in good growth condition with 90% confluence were trypsinized and prepared into a single cell suspension of 5 × 104 cells/mL with complete medium and inoculated onto adherent slides; cells were washed twice with PBS and labeled with 50nM MitoTracker™ Red (red), incubated for 20 min in the incubator. The cells were washed twice with PBS, the nuclei were labelled with 1 µg/ml Hoechst 33,342 working solution (blue), the cells were washed twice with PBS, the slides were fixed and sealed, and the mitochondria were observed by laser confocal microscopy and photographed.
qRT-PCR detection
Total RNA was extracted using the Trizol reagent (Thermo Fisher Scientific, USA). Reverse transcription was performed using a 5X All-In-One RT MasterMix (Applied Biological Materials, CAN) and RT-PCR was performed using EvaGreen Express 2×qPCR MasterMix-Low Rox (Applied Biological Materials, CAN). The ΔΔCt method was used for the calculation of relative changes in gene expression. The primer sequences for RT-PCR were listed in Table 1.
Western blot analysis
Total protein was extracted using RIPA buffer with protease inhibitors (Boster, USA). For immunoblot analysis, protein samples were resolved by SDS-PAGE and transferred onto PVDF membranes by Mini-PROTEAN Tetra system (Bio-Rad Laboratories, USA). PVDF membranes were blocked in 1×TBST containing 5% non-fat dry milk for 1 h and incubated with primary antibodies overnight at 4 °C. PVDF membranes were washed 3 times with TBST and incubated with HRP-conjugated secondary antibodies (Abcam, UK) for 1 h at room temperature. SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, USA) was used for visualization and imaging. Signals were detected and quantified using Image Lab 4.0 software (Bio-Rad Laboratories, USA). The primary antibodies were listed in Table 2.
Statistical analysis
Statistical analysis was performed using SPSS19.0. Data are expressed as mean ± SD. One-way analysis of variance (ANOVA) was used for statistical analysis of data within and between groups, followed by the least significant difference (LSD) test for multiple comparisons of group means. P < 0.05 was considered statistically significant.