Abstract
Background
Cellular immune responses are phenotypically and functionally perturbed during HIV-1 infection, with the majority of function restored upon antiretroviral therapy (ART). Despite ART, residual inflammation remains that can lead to HIV-related co-morbidities and mortality, indicating that ART does not fully restore normal immune cell function. Thus, understanding the dynamics of the immune cell landscape during HIV-1 infection and ART is critical to defining cellular dysfunction that occurs during HIV-1 infection and imprints during therapy.
Results
Here, we have applied single-cell transcriptome sequencing of peripheral blood immune cells from chronic untreated HIV-1 individuals, HIV-1-infected individuals receiving ART and HIV-1 negative individuals. We also applied single-cell transcriptome sequencing to a primary cell model of early HIV-1 infection using CD4+ T cells from healthy donors. We described changes in the transcriptome at high resolution that occurred during HIV-1 infection, and perturbations that remained during ART. We also determined transcriptional differences among T cells expressing HIV-1 transcripts that identified key regulators of HIV-1 infection that may serve as targets for future therapies to block HIV-1 infection.
Conclusions
This work identified key molecular pathways that are altered in immune cells during chronic HIV-1 infection that could remain despite therapy. We also identified key genes that are upregulated during early HIV-1 infection that provide insights on the mechanism of HIV-1 infection and could be targets for future therapy.
Similar content being viewed by others
Background
HIV-1 infection leads to progressive loss of CD4+ T cells that results in the development of acquired immunodeficiency syndrome (AIDS) [1, 2]. Initiation and adherent use of antiretroviral therapy (ART) can achieve viral suppression that decreases HIV-1 mortality and reduces the potential to transmit the virus to others [3,4,5]. In addition, the chronic inflammation caused by the virus has been linked to non-AIDS related morbidity and mortality making it clear that HIV-1 damages the host immune system and ART may only partially restore the damage and the level of activation of the immune system [6,7,8]. These observations coupled with the number of new HIV-1 infections that occur despite prevention and therapeutic efforts highlight the need to better understand the impact HIV-1 has on host immune cell function in order to understand infection and improve therapeutic outcomes.
Recently, high-throughput single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool to understand the transcriptional differences in populations of cells at single-cell resolution and has provided critical insights to understanding human disease pathogenesis. Single-cell approaches have been used to study HIV-1 replication and infection, the heterogeneity of the HIV-1 latent reservoir and the cellular response to latency reversal agents [9,10,11,12,13]. Many of these studies have focused on the effect of HIV-1 on CD4+ T cells and have identified key pathways that are important for the regulation of HIV-1 infection and latency. Study of non-CD4+ T cell populations has been limited, but investigators have demonstrated the complex interplay of innate and adaptive immunity in the regulation of the HIV-1 broadly neutralizing antibody responses and viremia during infection [14,15,16].
Here, we sequenced the transcriptomes of 81,235 peripheral blood immune cells from six untreated chronic HIV-1-infected individuals, three ART-treated HIV-1-infected individuals and three HIV-1 seronegative individuals. We also identified cell types and states that changed during infection and described changes in immune cells that remain during ART. Finally, we used the scRNA-seq approach in a primary cell model of acute infection with HIV-1 to identify gene expression changes that occur in cells that express HIV-1 compared to those without HIV-1 transcript expression.
Results
scRNA-seq identified distinct immune cell populations during HIV-1 infection
To determine the host immune cell transcriptional changes during HIV-1 infection and ART, we performed scRNA-seq of peripheral blood mononuclear cells (PBMCs) from six untreated chronically HIV-1-infected (untreated), three ART-treated HIV-1-infected (treated) and three HIV-1 seronegative control (seronegative) participants using the 10 × Genomics scRNA-seq platform (Table 1). The HIV-1-infected individuals on ART had undetectable viral load, but were only on ART for less than one year which could allow for residual viral replication compared to individuals on ART for longer periods. In order to normalize the data and to filter out low-quality cells, we utilized the CellRanger aggregation pipeline which normalized for effective sequencing depth of each sample by subsampling the reads to the sample with the lowest depth. Next, we excluded cells that had greater than 20% of the genes that were mitochondrial genes (Additional file 1: Fig. S1A). Finally, to correct for any batch effects we used the Seurat analysis pipeline Multi CCA method to regress out cell–cell variation in gene expression in order to control for technical variation. This resulted in the elimination of 994 cells from the final dataset. Among all individuals, we determined the transcriptomes of 81,297 single cells detecting a median of 951 genes per cell (Fig. 1A; Additional file 1: Table S1).
scRNA-seq of peripheral immune cells identified distinct cellular populations. A Schematic of scRNA-seq experimental procedure using peripheral blood mononuclear cells (PBMC) from 6 untreated chronic HIV-1-infected individuals, 3 ART-treated chronic HIV-1-infected individuals and 3 HIV-1 seronegative individuals resulting in the analysis of 81,235 total PBMC cells. B UMAP analysis of all 81,235 cells and graph-based clustering of cell populations resulted in the identification of 23 transcriptionally distinct clusters. Individual cells are colored by cluster identity. C UMAP analysis with cell colored and labeled by cell identify determined by Azimuth reference. D UMAP analysis with individual cells colored by sample group. E Bar graph showing the fraction of total single cells in each Azimuth defined cell type by group
We first used an unsupervised analysis approach and performed dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) and graph-based clustering to visualize and identify 23 transcriptionally distinct cell clusters in the total single cell dataset (Fig. 1B). To define broad cell types contained in each cluster we used the SingleR reference-based map** to assign cell type identities to each cluster using both the Immune cell expression and Monaco Immune databases (Additional file 1: Table S2). In parallel, we determined the pattern of expression of transcripts that define immune cell identity to further classify the cell clusters. We determined the expression of CD14 expressed in monocytes; CD3D, CD4, CD8A expressed in T cell populations; KLRC1 expressed in natural killer (NK) cell populations; and CD79A expressed in B cell populations (Additional file 1: Fig. S2A). We then identified the genes that were uniquely upregulated in each cell cluster, and used the top three upregulated genes in each cluster to generate a heatmap of gene expression across the clusters (Fig. 1D; Additional file 2: Table S3).
Next, we used a supervised analysis approach to identify cell types but also more specific cell states using the Azimuth reference-based map** pipeline using refence PBMC datasets. This identified naïve, memory, cytotoxic and regulatory populations of both CD4 and CD8 T cells, natural killer (NK) cell, B cells, monocyte, dendritic cells and small populations or platelet and erythroid cells (Fig. 1C). This analysis recapitulated many of the expected immune cell lineages present in PBMCs.
We identified cells by the HIV group and found that cells from HIV-1 infection (treated and untreated) had overlap** populations with the HIV-1 seronegative samples indicating overall similar transcriptomes of certain cellular populations, as well as clusters of cells that displayed a unique transcriptome (Fig. 1D; Additional file 1: Fig. S2). We then determined the fraction of cells in each Azimuth-identified cell type that was had a frequency above 1% of the total cells (Fig. 1E). The most abundant cell types in all groups were CD4 T central memory, CD8 T effector memory and CD14 monocytes. Individuals with HIV-1 infection (treated and untreated) had increased frequency of CD8 T effector memory cells (29.2%, ART HIV-1; 28.6%, Untreated HIV-1) compared to seronegative individuals (14.2%; Fig. 1E). Conversely, HIV-infected individuals had reduced frequencies of CD4 T central memory cells (20.0%, ART HIV-1; 14.0%, Untreated HIV-1) compared to seronegative individuals (27.2%; Fig. 1E). Individuals with treated or untreated HIV-1 infection also had increased frequencies of CD14 (13.2% ART HIV-1; 19.3%, Untreated HIV-1) and CD16 (5.5%, ART HIV-1; 7.2%, Untreated HIV-1) monocyte populations compared to the CD14 (11.5%) and CD16 (3.6%) monocyte populations in seronegative individuals (Fig. 1E). The reduction in CD4 T cell and increase in CD8 T and monocyte populations were also evident when the fraction of cells within each unsupervised defined cluster (Additional file 1: Fig. S2C). Despite ART treatment and no detectable HIV-1 viral load, transcriptionally unique populations of immune cells were still present in the treated group especially among monocytes and T cells (clusters 4, 6, 12, 16), indicating that ART did not fully restore the immune cell transcriptional landscape after infection (Figs. 1E; Additional file 1: Fig. S2). Cluster 6 represented over 20% of the cells in the treated group and contained a population of regulatory T cells. Clusters 12 and 16 were also nearly exclusively made up of cells in the treated group and represented populations of innate like cells (ILC) and CD16 monocytes, respectively (Additional file 1: Fig. S2C; Additional file 2: Table S3). These data revealed that HIV infection perturbed immune cell identity and that short-term treatment to undetectable HIV-1 viral load did not fully restore the cellular phenotypes.
Identification of differentially regulated genes in immune cells from HIV-1-infected and seronegative individuals
We next determined the genes that were differentially expressed in cells from the untreated HIV-1-infected individuals compared with the seronegative individuals. We found 96 genes that were upregulated and 45 genes that were downregulated in the cells from untreated HIV-1-infected individuals (Fig. 2A; Additional file 3: Table S4). The top seven upregulated genes are all associated with interferon response (S100A8/9, MT2A, IFITM3, IFI6, ISG15) [17], while the top downregulated gene ZNF90 is a transcription factor with unknown roles in immune cells followed by the genes CXCR4, IL7R and BTG1, which are key regulators of lymphocyte proliferation and motility.
Differentially expressed genes in untreated chronic and ART treated HIV-1-infected compared with HIV-1 seronegative cells. A–B Volcano plot of genes that are significantly (P ≤ 0.05; Wilcoxon rank sum test, Bonferroni adjusted) different when comparing (A) Untreated HIV-1 vs. Seronegative and B ART HIV-1 vs. Seronegative group cells. X-axis displays log2 fold-change of each significant gene. C–D Gene Set Enrichment Analysis (GSEA) of the top enriched biological pathways in genes significantly altered in C untreated HIV-1 or D DART HIV-1. Pathways highlighted in red are present in both groups. GSEA enrichment plot for the Hallmark_Inflammatory_Response feature is shown below each graph
Next, we identified transcripts that were differentially expressed in cells from HIV-1-treated individuals compared with the HIV-1 seronegative individuals to determine genes that are changed during HIV-1 infection despite ART control of viral load. We found 83 genes upregulated and 31 genes downregulated in cells from the HIV-1-treated individuals compared to seronegative individuals (Fig. 2B; Additional file 4: Table S5). CCL4 was the highest upregulated transcript in cells from treated individuals. The CCL4 gene encodes the protein MIP-1b that can bind HIV-1 co-receptor CCR5 and inhibit infection [18]. The next three upregulated genes in treated individuals were DNAJB1, which is a part of the HSP40 complex that has been shown to play a role in anti-inflammatory processes in autoimmunity [19]; and RGS1 and SOD1, which are important for antiviral immune responses [20]. The top downregulated transcripts in cells from treated individuals were the transcription factor ZNF90; NHSL2, which has unknown function; and IL7R, which has been shown to play a role in CD4 T cell loss during HIV-1 infection [21].
We performed Gene Set Enrichment Analysis (GSEA) using the differentially expressed transcripts in untreated HIV-1 or ART-treated HIV-1 compared to seronegative individuals and plotted the hallmark pathways that were significantly enriched (Fig. 2C, D). There were 10 hallmarks that had significant FDR-corrected q-values for the untreated HIV-1 compared to seronegative controls and 15 hallmarks that were significant in the ART-treated individuals (Additional file 1: Table S6 and S7). Both gene sets were enriched for pathways involved in inflammatory responses, complement immune responses and interferon responses (Fig. 2C, D). There were also several enriched pathways involved in metabolism and cell death. The Hallmark Inflammatory Response was significantly enriched in both HIV-1-infected groups (Fig. 2C, D). These data demonstrated that genes associated with the interferon response are upregulated in cells during HIV-1-infection, and key immune genes important for inflammatory processes and controlling virus replication, such as CCL4, remain changed even during ART therapy.
T cells from ART-treated individuals have increased regulatory transcripts
We selected and performed graph-based clustering of cells that identified as T cells and NK cells from the larger dataset to determine higher resolution differences in cellular clusters (Clusters 0, 1, 2, 3, 4, 6, 10, 12, 13, 14, 19, 21; Fig. 1). We identified 17 new distinct transcriptional clusters within this subset of cells (Fig. 3A). We next determined the genes that were upregulated within each cluster of cells, and generated a heatmap of the top five genes that defined each cluster to further classify the identity of each cell cluster (Additional file 1: Fig. S3A; Additional file 5: Table S8). We also determined the expression of nine specific lineage-defining transcripts in order to further classify the cell clusters (Additional file 1: Fig. S3B). CD3D encodes for CD3 which identified T cell populations and the T cells were further subdivided into CD4+ and CD8+ T cells using CD4 and CD8A expression, respectively. This analysis identified a single major cluster of CD4 T cells, but multiple CD8 T cell clusters that were transcriptionally distinct (Additional file 1: Fig. S3B). We found that the T cells were segregated by naïve T cell marker CCR7 as well as a marker of T cell activation MKI67. There were also smaller populations of T cells with the regulatory marker FOXP3 and exhaustion marker PD-1 (PDCD1; Additional file 1: Fig. S3B). We also identified two major clusters of cells that lacked T cell markers (cluster 11 and 13). Cluster 11 was further classified as NK cells due to expression of granzyme B and CD16 transcripts (GZMB and FCER1G) and cluster 13 had markers of monocyte populations (CD16, FCER1G; lysozyme, LYZ; Additional file 1: Fig. S3). In parallel, we utilized the reference-based cell identify map** approach Azimuth to identify cell populations. This revealed similar cellular populations of CD4 and CD8 naïve and memory populations, T regulatory cells, and NK cells on the major group of single-cells on the UMAP. There were also smaller segregated populations of proliferating cell types, and platelets, B cells, monocytes and dendritic cells (Fig. 3B).
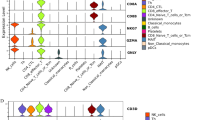
Proinflammatory signatures in T cells despite therapy. A UMAP plot identifying T cell clusters that were isolated and reclustered for more high-definition analysis revealing multiple cellular clusters. B UMAP analysis with cell colored and labeled by cell identify determined by Azimuth reference. C UMAP plots of T cell clusters with cells labeled and colored by experimental group. D Bar graph showing the fraction of total single cells in each Azimuth defined cell type by group. E–F Differentially expressed genes in Chronic or ART-treated HIV-1-infection compared to cells from seronegative individuals in E CD4 T cells or F CD8 T cells. Each dot represents a gene and Bonferroni adjusted p value displayed on Y-axis and the Log2 Fold change of expression displayed on the X-axis
Cells from untreated HIV-1 and seronegative individuals had many overlap** clusters, while the cells from the ART-treated individuals had more unique clusters of T cells (Fig. 3C). Similar to what was observed when analyzing the entire PBMC dataset, there were smaller frequencies of CD4 TCM and increased frequencies of CD8 TEM cell populations in the cells from the untreated or treated HIV-1 infected individuals compared to the cells from the seronegative individuals (Fig. 3D). There was a shift in the T cell populations for ART-treated HIV-1 individuals that were made up of clusters 4, 5 and 10 and represented mostly CD4 cytotoxic T lymphocytes (CTL; Fig. 3A–D). These cells represented 6.14% of the NK/T cells from ART-treated individuals compared to 2.46% and 1.67% of the untreated HIV-1 and seronegative groups, respectively (Fig. 3D). Notably, these clusters had upregulation of two genes that have been shown to play a role in the development of regulatory T cells NFKBIA and FTH1 [22] (Additional file 1: Fig. S3A).
Next we performed differential gene expression comparisons of the CD4+ and CD8+ T cell populations. CD4+ and CD8+ T cells (CD4 and CD8A-expressing cells, respectively) from untreated HIV-1 individuals had upregulation of interferon response genes (IFI6, MT2A, S100A8, S100A9) and downregulation of the transcription factor ZNF90 and IL7R that encodes CD127, important for T cell differentiation [23], when compared to cells from seronegative donors (Additional file 6: Table S9 and Additional file 7: Table S10). When comparing differentially expressed genes in cells from ART individuals and seronegative, we found that CD4+ T cells had upregulation of chemokines and IL1B, which contribute to the proinflammatory state (Fig. 3E; Additional file 8: Table S11). In CD8+ T cells from untreated HIV-1-infected compared to seronegative individuals, there is upregulation of LGALS1 that encodes galectin-1 that has been shown to induce apoptosis in effector T cells and increase immunoregulatory roles of T cells [24] (Fig. 3F; Additional file 9: Table S12). CD8+ T cells had upregulation of DUSP2 and RGS1 that are important for T cell inflammation and cell migration [25] (Fig. 3F; Additional file 9: Table S12). We observed a similar downregulation of ZNF90 and IL7R when we compared ART to seronegative cells as when we compared untreated HIV cells were compared to seronegative, indicating these transcripts are changed in T cells during untreated or treated HIV-1 infection (Fig. 3E, F). These data demonstrated that CD4+ and CD8+ T cells remain in a proinflammatory state despite ART.
Increased expression of T cell activation and long-noncoding RNA transcripts regulate HIV-1 infection
To identify genes that are critical for regulating CD4+ T cell resistance or sensitivity to initial HIV-1 infection, we used scRNA-seq in a primary CD4+ T cell model of HIV-1 infection. CD4+ T cells were enriched from the PBMCs of two healthy HIV-1 uninfected donors and stimulated for 72 h with recombinant human IL-2 and antibodies directed against CD3 and CD28. After stimulation, the cells were infected with a full-length primary HIV-1 infectious molecular clone (CH058; [26]) for 24 h. Cells were harvested and subjected to scRNA-seq (Fig. 4A). We analyzed 7359 single T cells from the two donors after filtering for poor quality cells (Additional file 1: Fig. S1B). The cells from both donors clustered similarly using UMAP (Fig. 4B). Annotation of reference-based cell identities using Azimuth revealed that most cells were CD4 proliferating cells, with small populations of cells with CD4 TCM identity (Fig. 4C). There were also some cells that had gene expression profiles that matched CD8 naïve, CD8 and NK proliferating cells (Fig. 4C). Expression of HIV-1 gag transcripts was detected in 2005 of the cells, indicating that after 24 h of virus exposure approximately 27% of the cells had active viral transcription (Fig. 4D). In addition to HIV-1 gag transcript, we identified 12 genes differentially expressed in cells with HIV-1 transcripts compared with HIV-1 negative cells (Fig. 4E, F). Two long-noncoding RNAs (NEAT1 and MALAT1) were upregulated as were other markers of T cell activation and interferon signaling (CD69, CCL3, TNFRSF18, GZMB, GZMA, PRDM1, IFNG; Fig. 4E, F). The three downregulated transcripts had variable function in cell metabolism, transcription and immune signaling (HSP90AA1, EIF5A, CYP1B1; Fig. 4F). Notably, the top three genes that correlated (Pearson) with HIV-1 gag expression levels were the two long-noncoding RNAs NEAT1 and MALAT1, and PRDM1, which encodes the transcriptional regulator BLIMP1 (Fig. 4G; Additional file 10: Table S13). All three of these genes have been implicated in controlling HIV-1 virus transcription and latency [27,28,29]. Among the genes that anti-correlated with gag expression were members of the SR protein splicing factor family (SRSF2, SRSF3, SRSF7) that have been shown to be important for HIV-1 RNA splicing [30] (Fig. 4G; Additional file 10: Table S13). These data show that the two long-noncoding RNAs NEAT1 and MALAT1, SR protein splicing factors, and T cell activation status are critical for acute HIV-1 infection and viral gene transcription. Future studies will be required to determine if manipulation of these genes affects sensitivity to HIV-1 infection will be required.
CD4 T cell activation status and long-noncoding RNAs regulate early HIV-1 infection A overview of primary cell model of HIV-1 infection and scRNA-seq. B UMAP plot of scRNA-seq of T cell data from two independent healthy individuals. Each point represents a single cell. C UMAP analysis with cell colored and labeled by cell identify determined by Azimuth reference. D UMAP plot displaying HIV-1 gag gene expression in the individual cells. E UMAP plots showing expression of transcripts that were significantly upregulated in T cells that had expression of HIV-1 gag compared to cells that lacked expression. F Transcripts that were significantly changed in T cells that had expression of HIV-1 gag compared to cells that laced detectable expression. G Top genes that correlated and anti-correlated (r, Pearson) with HIV-1 gag expression in all cells
Discussion
In this study, we have applied single-cell transcriptome sequencing to map at high resolution the alterations and cellular dynamics associated with HIV-1 infection and therapy. Chronic infection with HIV-1 causes progressive loss of immune cell function that leads to impaired immunity [31]. This immune dysfunction is partially restored through antiretroviral therapy, but the molecular mechanisms of the development and restoration of immune cell exhaustion are not fully understood, nor the immune cell perturbations that remain despite therapy [32, 33]. Immune activation during HIV-1 infection is not limited to the target CD4+ T cells but engages a range of molecular and cellular processes in both the innate and adaptive arms of the immune system [34]. Using single-cell transcriptomics, we were able to identify immune cell perturbations in subsets of immune cells that were changed in individuals with HIV-1 and some of which persisted in individuals on therapy.
Confirming previously published studies, HIV-1 infection exhibited a proinflammatory immune state in all immune cell types we analyzed, with evidence of upregulation of interferon response genes [13, 35, 36]. When comparing immune cells from ART-treated HIV-1-infected individuals, we found higher frequencies of activated B cells and proinflammatory CD4+ and CD8+ T cell populations than in seronegative controls. These data demonstrated that even when viral load is below the limit of detection, changes in immune cells may persist. The shift of immune cells to a more proinflammatory state could contribute to the comorbidities observed in HIV-1 individuals despite ART [6, 37]. It will be important to study individuals at variable times after therapy initiation to determine if and when immune cell states return to pre-infection levels.
Finally, we applied this technology to a primary CD4+ T cell model of early HIV-1 infection to identify genes that directly contribute to sensitivity or resistance to HIV-1 infection. We identified two long-noncoding RNAs, NEAT1 and MALAT1 that correlated with HIV-1 transcript levels. These two lncRNAs have previously been shown to regulate T cell activation and control HIV-1 transcription, but their role in regulating the cell sensitivity to HIV-1 infection has not been determined [28, 29, 38]. Other studies using single-cell genomic approaches have determined that HIV-1-infected cells undergo clonal expansion due to antigenic or cytokine stimulation that help with seeding the latent reservoir that are increased over time [39]. Understanding the clonal T cell dynamics during infection and during therapy will be important for future studies. Moreover, studies have identified that the proviruses that make up the latent reservoir are enriched in sequences that are present near ART initiation [40, 41]. This could suggest regulation of long-term persistence of HIV-infected cells. The genes identified in our primary model of acute infection could indicate that there are cellular pathways that restrict HIV replication and establishment of persisting infected cells for long-term. These genes, along with RNA splicing factors identified in our analysis, may be targeted for future therapies to block HIV-1 infection or control viral transcription in HIV-1 cure strategies.
There are limitations to our study, the major one is that small exploratory study size. We know that demographic and other host factors such as age, sex, race/ethnicity, other health conditions all can effect host immune cell profiles. Moreover, the comparison of ART drug regimens and individuals with different duration of ART would be informative that was not addressed here. Larger studies that control for infection history ART class and duration as well as account for the impact of demographics on immune cell profiles will be required to validate any discoveries. Transcripts that have lower expression in the single-cell datasets could also be critical for HIV immune regulation but could have the impact reduced due to low and variable detection. Improved scRNA-seq sensitivity or targeted approaches on selected low-abundant transcripts could highlight the significance of these genes. Another limitation is we studied the immune cells in only the peripheral blood. Determining how HIV-1 infection and treatment influences cells in other tissues such as the gut and lymphoid organs will be critical in understanding how the virus impacts immune cells in these critical tissue environments.
Conclusions
In summary, our study demonstrates the ability of scRNA-seq to overcome the obstacles of the complexity and heterogeneity of the immune system. Using this approach, we generated a high-resolution atlas of the molecular changes that occur in blood immune cells during HIV-1 infection and ART treatment, uncovering novel genes and inflammatory pathways that may be important for restoring immune homeostasis in individuals with HIV-1 infection. Integration of the HIV-1 viral genes to identify infected cells and determine transcriptome changes that occur during infection of the target cells provides a framework to study HIV-1 and other viral infections such as influenza, SARS-CoV-2 and measles that are known to perturb the gene expression of host cells. Taken together, using this approach in other infection and disease contexts will reveal unique and overlap** immune pathways that could be targeted with drugs or treatment in the future.
Methods
Experimental model and subject details
Deidentified peripheral blood mononuclear cell (PBMC) samples were used from existing cohorts of HIV-1-infected and seronegative individuals stored at the Duke Human Vaccine Institute that were approved by the Duke Medicine Institutional Review Boards as well as the ethics boards of the local sites. Individuals were mostly male and of African descent (Table 1). These individuals were a part of a large HIV-1 clinical study that received longitudinal tissue sample collection. Whole venous blood was drawn and then fractionated into PBMCs and cryopreserved. This was an exploratory study, and the selected participants were not statistically matched for external factors such as age, sex, viral load or ethnicity. Future studies should be performed to confirm these cofounders. Cells from six untreated HIV-infected, three ART-treated HIV-1-infected (HIV-1 viral load below the limit of detection), and three HIV-1 seronegative (control) individuals were used to perform single-cell RNA sequencing studies (10× Genomics; Table 1).
Primary cell model of HIV-1 infection
PBMCs from two different HIV-1 seronegative individuals (not included in other analysis) were thawed and T cells were activated by stimulation with CD3 (clone OKT3; eBioscience, 150 ng/mL) and CD28 (clone CD28.2; BD Biosciences, 150 ng/mL) antibodies in RPMI media supplemented with 20% FBS and recombinant human IL-2 (NIH AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health, 30 U/mL). Activation was allowed to proceed for 72 h at 37 °C 5% CO2. CD4+ T cells were then enriched by removal of CD8+ T cells using magnetic CD8 microbeads (Miltenyi Biotec, Germany). Cells were then infected with full-length HIV-1 infectious molecular clone virus derived from an infected individual CH058 [26] by spinoculation at 1200×g for 2 h at room temperature [42]. Infected cells were incubated in RPMI media supplemented with 20% FBS and 30 U/mL IL-2 for 24 h. Cells were then harvested and washed with PBS for scRNA-seq.
scRNA-seq library construction and sequencing
PBMCs were thawed, washed and placed in single-cell suspensions with PBS + 0.04% BSA. Single-cells after activation and 24 h infection from the primary cell model of HIV-1 infection were also washed and also placed in single-cell suspension with PBS + 0.04% BSA. Cellular suspensions were loaded on a GemCode Single-Cell instrument (10× Genomics) to generate single-cell beads in emulsion. Single-cell RNA-seq libraries were then prepared using a GemCode Single Cell 3’ Gel bead and library kit version 2 (10× Genomics). Single-cell barcoded cDNA libraries were quantified by quantitative PCR (Kappa Biosystems) and sequenced on an Illumina NextSeq 500 [9, 14, 43, 44]. Read lengths were 26 bp for read 1, 8 bp i7 index, and 98 bp read 2. Cells were sequenced to greater than 50,000 reads per cell.
scRNA-seq map** and analysis
After sequencing, the Cell Ranger Single Cell Software Suite (version 2.1.1) was used to generate sequencing fastq files and to perform sample de-multiplexing, barcode processing, reference alignment and single-cell 3’ gene counting [45]. Reads were aligned to the human genome (GRCH38) combined with the consensus sequence of HIV-1 clade C gag derived from viruses in the HIV Sequence Database (https://www.hiv.lanl.gov). Clade C was selected due to it being the predominant circulating HIV-1 strain in southern Africa where most of the individuals originate. Samples were aggregated using the CellRanger Aggr function to create a single matrix of cell barcodes and gene counts for the groups. During the process each library is normalized for mapped sequencing depth. Reads are subsampled from higher-depth libraries until they all have an equal number of reads per cell that are confidently mapped to the transcriptome in order to control for variation in the number of reads per sample (sequencing depth). Finally, to correct for any batch effects we used the Seurat analysis pipeline Multi CCA method to regress out cell–cell variation in gene expression in order to control for technical variation. The union of variable genes across all individual samples are utilized to renormalize the data.
Matrices of cell barcodes and gene counts generated by Cell Ranger were loaded into Seurat R package (v3.2.3) for graph-based cell clustering, dimensionality reduction and data visualization [46,47,48]. We filtered low quality cells that had lower than 200 expressed transcripts and percentage of mitochondrial genes expressed greater than 20% and for the primary cell model we reduced this threshold to greater than 10% mitochondrial genes. We included up to 45 PCA dimensions for the PBMCs and 48 PCA for the primary cell model for downstream graph-based clustering and UMAP visualization. All other parameters we followed the default Seurat recommendations. SingleR (v. 1.0.5) was utilized to assist with immune cell type identification [49]. Supervised identification of cell identity and states were performed using Azimuth [50]. Cells were annotated using the RunAzimuth, a Seurat object was provided as a query, function available on azimuth package in R using default settings. The Human-PBMC dataset was used as the reference dataset for Azimuth [48]. The reported cell type annotations are based on “celltype.I2” Azimuth output. Differentially expressed genes between cell clusters or groups were determined using Seurat by the Wilcoxon rank sum test. Genes that correlated with HIV-1 transcript expression were calculated by Pearson correlation and corrected for multiple comparisons using Bonferroni. Graphs and plots were generated using the Seurat and ggplot2 (v3.3.3) R packages and Graphpad Prism version 8.
GSEA v4.1.0 was used for gene set enrichment analysis. For the top 2000 variable genes, raw counts were linearly transformed (using ScaleData function available on Seurat Package) and averaged to generate expression dataset. Next, phenotype file was prepared based on group information for the samples. Hallmark gene sets (v7.2) and Human_HGNV_ID_MiSigDB (v7.2) chip hosted on the GSEA-MSigDB file servers were used as Gene sets and Chip annotation file respectively. Genes weren’t collapsed and “gene_set” parameter was used for permutation. While rest of the options were run with default settings.
Availability of data and materials
The single-cell RNA seq unprocessed reads have been deposited in the NCBI SRA database under the BioProject ID: PRJNA681021.
Code availability
Code and other processed file formats are available from corresponding authors upon reasonable request.
References
Doitsh G, Greene WC. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe. 2016;19(3):280–91.
Klatzmann D, Barre-Sinoussi F, Nugeyre MT, Danquet C, Vilmer E, Griscelli C, et al. Selective tropism of lymphadenopathy associated virus (LAV) for helper-inducer T lymphocytes. Science. 1984;225(4657):59–63.
Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med. 2016;375(9):830–9.
Rodger AJ, Lodwick R, Schechter M, Deeks S, Amin J, Gilson R, et al. Mortality in well controlled HIV in the continuous antiretroviral therapy arms of the SMART and ESPRIT trials compared with the general population. AIDS. 2013;27(6):973–9.
Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–60.
Zicari S, Sessa L, Cotugno N, Ruggiero A, Morrocchi E, Concato C, et al. Immune activation, inflammation, and non-AIDS co-morbidities in HIV-infected patients under long-term ART. Viruses. 2019;11(3):66.
Peterson TE, Baker JV. Assessing inflammation and its role in comorbidities among persons living with HIV. Curr Opin Infect Dis. 2019;32(1):8–15.
Gay CL, Mayo AJ, Mfalila CK, Chu H, Barry AC, Kuruc JD, et al. Efficacy of NNRTI-based antiretroviral therapy initiated during acute HIV infection. AIDS. 2011;25(7):941–9.
Bradley T, Ferrari G, Haynes BF, Margolis DM, Browne EP. Single-cell analysis of quiescent HIV infection reveals host transcriptional profiles that regulate proviral latency. Cell Rep. 2018;25(1):107–17e3.
Cohn LB, da Silva IT, Valieris R, Huang AS, Lorenzi JCC, Cohen YZ, et al. Clonal CD4(+) T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat Med. 2018;24(5):604–9.
Golumbeanu M, Cristinelli S, Rato S, Munoz M, Cavassini M, Beerenwinkel N, et al. Single-cell RNA-Seq reveals transcriptional heterogeneity in latent and reactivated HIV-infected cells. Cell Rep. 2018;23(4):942–50.
Rato S, Rausell A, Munoz M, Telenti A, Ciuffi A. Single-cell analysis identifies cellular markers of the HIV permissive cell. PLoS Pathog. 2017;13(10): e1006678.
Kazer SW, Aicher TP, Muema DM, Carroll SL, Ordovas-Montanes J, Miao VN, et al. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat Med. 2020;26(4):511–8.
Bradley T, Peppa D, Pedroza-Pacheco I, Li D, Cain DW, Henao R, et al. RAB11FIP5 expression and altered natural killer cell function are associated with induction of HIV broadly neutralizing antibody responses. Cell. 2018;175(2):387–99e17.
Luban J. Innate immune sensing of HIV-1 by dendritic cells. Cell Host Microbe. 2012;12(4):408–18.
Scully E, Alter G. NK cells in HIV disease. Curr HIV/AIDS Rep. 2016;13(2):85–94.
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–6.
Abdelwahab SF, Cocchi F, Bagley KC, Kamin-Lewis R, Gallo RC, DeVico A, et al. HIV-1-suppressive factors are secreted by CD4+ T cells during primary immune responses. Proc Natl Acad Sci USA. 2003;100(25):15006–10.
Kotlarz A, Tukaj S, Krzewski K, Brycka E, Lipinska B. Human Hsp40 proteins, DNAJA1 and DNAJA2, as potential targets of the immune response triggered by bacterial DnaJ in rheumatoid arthritis. Cell Stress Chaperones. 2013;18(5):653–9.
Kane M, Zang TM, Rihn SJ, Zhang F, Kueck T, Alim M, et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20(3):392–405.
Bazdar DA, Kalinowska M, Sieg SF. Interleukin-7 receptor signaling is deficient in CD4+ T cells from HIV-infected persons and is inversely associated with aging. J Infect Dis. 2009;199(7):1019–28.
Cuadrado E, van den Biggelaar M, de Kivit S, Chen YY, Slot M, Doubal I, et al. Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity. 2018;48(5):1046–59e6.
Paiardini M, Cervasi B, Albrecht H, Muthukumar A, Dunham R, Gordon S, et al. Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J Immunol. 2005;174(5):2900–9.
Cedeno-Laurent F, Dimitroff CJ. Galectin-1 research in T cell immunity: past, present and future. Clin Immunol. 2012;142(2):107–16.
Lang R, Raffi FAM. Dual-specificity phosphatases in immunity and infection: an update. Int J Mol Sci. 2019;20(11):66.
Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol. 2012;86(5):2715–28.
Kaczmarek Michaels K, Natarajan M, Euler Z, Alter G, Viglianti G, Henderson AJ. Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J Immunol. 2015;194(7):3267–74.
Liu H, Hu PW, Couturier J, Lewis DE, Rice AP. HIV-1 replication in CD4(+) T cells exploits the down-regulation of antiviral NEAT1 long non-coding RNAs following T cell activation. Virology. 2018;522:193–8.
Qu D, Sun WW, Li L, Ma L, Sun L, ** X, et al. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47(6):3013–27.
Mahiet C, Swanson CM. Control of HIV-1 gene expression by SR proteins. Biochem Soc Trans. 2016;44(5):1417–25.
Ruegg CL, Engleman EG. Impaired immunity in AIDS. The mechanisms responsible and their potential reversal by antiviral therapy. Ann NY Acad Sci. 1990;616:307–17.
Bohler T, Walcher J, Holzl-Wenig G, Geiss M, Buchholz B, Linde R, et al. Early effects of antiretroviral combination therapy on activation, apoptosis and regeneration of T cells in HIV-1-infected children and adolescents. AIDS. 1999;13(7):779–89.
Korencak M, Byrne M, Richter E, Schultz BT, Juszczak P, Ake JA, et al. Effect of HIV infection and antiretroviral therapy on immune cellular functions. JCI Insight. 2019;4(12):66.
Soudeyns H, Pantaleo G. The moving target: mechanisms of HIV persistence during primary infection. Immunol Today. 1999;20(10):446–50.
Nasr N, Alshehri AA, Wright TK, Shahid M, Heiner BM, Harman AN, et al. Mechanism of interferon-stimulated gene induction in HIV-1-infected macrophages. J Virol. 2017;91(20):66.
Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010;7:54.
Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39(4):633–45.
Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4(1):e00596–12.
Collora JA, Liu R, Pinto-Santini D, Ravindra N, Ganoza C, Lama JR, et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity. 2022;55(6):1013–31e7.
Abrahams MR, Joseph SB, Garrett N, Tyers L, Moeser M, Archin N, et al. The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci Transl Med. 2019;11(513):66.
Brodin J, Zanini F, Thebo L, Lanz C, Bratt G, Neher RA, et al. Establishment and stability of the latent HIV-1 DNA reservoir. Elife. 2016;5:66.
O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–80.
Bradley T, Kuraoka M, Yeh CH, Tian M, Chen H, Cain DW, et al. Immune checkpoint modulation enhances HIV-1 antibody induction. Nat Commun. 2020;11(1):948.
Han Q, Bradley T, Williams WB, Cain DW, Montefiori DC, Saunders KO, et al. Neonatal rhesus macaques have distinct immune cell transcriptional profiles following HIV envelope immunization. cell rep. 2020;30(5):1553–69e6.
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049.
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–14.
Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902e21.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, III, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–87e29.
Acknowledgements
The authors thank Caroline Cockrell, Thad Gurley and M. Anthony Moody for assistance with acquiring specimens from the Duke Human Vaccine Institute Accessioning Unit. The Duke Human Vaccine Institute Viral Genetics and Analysis Core for assistance with sequencing.
Funding
Research reported in this publication was supported by the NIH NIAID and the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID, UM1-AI100645), Duke Consortia for HIV/AIDS Vaccine Development (CHAVD, UM1-AI031308), and NIH NIAID project Grant R01-AI147778 (T.B.).
Author information
Authors and Affiliations
Contributions
JP performed investigation, data analysis and writing; SK performed data analysis; RWE performed investigation and data analysis; BH performed investigation and project administration; GF, BFH and TB performed investigation, conceptualization, data curation, funding acquisition, supervision and writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Deidentified peripheral blood mononuclear cell (PBMC) samples were used from existing cohorts of HIV-1-infected and seronegative individuals stored at the Duke Human Vaccine Institute that were approved by the Duke Medicine Institutional Review Boards as well as the ethics boards of the local sites (Duke IRB Pro00039127 and Pro00007562). Participants were originally enrolled in the study and written informed consent was obtained from all subjects and/or their legal guardian(s). All methods were carried out in accordance with the relevant guidelines and regulations of the Duke Medicine Institutional Review Boards as well as the ethics boards of the local sites in accordance with the Declaration of Helsinki. All experimental protocols reported in this manuscript with the human samples were reviewed and approved by the Duke institutional review board.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Table S1
: Individual sample scRNA-seq results. Table S2 Cell cluster computational inference of cell type. Table S6 GSEA enrichment of untreated HIV-1 compared to control. Table S7 GSEA enrichment of ART-treated HIV-1 compared to control. Fig. S1. scRNA-seq individual cell QC data. Fig. S2. Determining cell types and genes in each cluster. Fig. S3. NK and T cell reclustering.
Additional file 2. Table S3
: Genes upregulated in each cluster in scRNA-seq total PBMC.
Additional file 3. Table S4
: Genes differentially expressed between HIV-1-infected compared to seronegative.
Additional file 4. Table S5
: Genes differentially expressed between HIV-1-treated compared to seronegative.
Additional file 5. Table S8
: Genes upregulated in each cluster in scRNA-seq T and NK cells.
Additional file 6. Table S9
: Genes differentially expressed between HIV-1-infected compared to seronegative CD4+ T cells.
Additional file 7. Table S10
: Genes differentially expressed between HIV-1-infected compared to seronegative CD8+ T cells.
Additional file 8. Table S11
: Genes differentially expressed between HIV-1-treated compared to seronegative CD4+ T cells.
Additional file 9. Table S12
: Genes differentially expressed between HIV-1-treated compared to seronegative CD8+ T cells.
Additional file 10. Table S13
: Genes correlated or anti-correlated with HIV-1 gag transcript expression.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pollara, J., Khanal, S., Edwards, R.W. et al. Single-cell analysis of immune cell transcriptome during HIV-1 infection and therapy. BMC Immunol 23, 48 (2022). https://doi.org/10.1186/s12865-022-00523-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12865-022-00523-2