Abstract
Background
Spermatogenesis is an intricate process regulated by a finely organized network. The orange-spotted grouper (Epinephelus coioides) is a protogynous hermaphroditic fish, but the regulatory mechanism of its spermatogenesis is not well-understood. In the present study, transcriptome sequencing of the male germ cells isolated from orange-spotted grouper was performed to explore the molecular mechanism underlying spermatogenesis.
Results
In this study, the orange-spotted grouper was induced to change sex from female to male by 17alpha-methyltestosterone (MT) implantation. During the spermatogenesis, male germ cells (spermatogonia, spermatocytes, spermatids, and spermatozoa) were isolated by laser capture microdissection. Transcriptomic analysis for the isolated cells was performed. A total of 244,984,338 clean reads were generated from four cDNA libraries. Real-time PCR results of 13 genes related to sex differentiation and hormone metabolism indicated that transcriptome data are reliable. RNA-seq data showed that the female-related genes and genes involved in hormone metabolism were highly expressed in spermatogonia and spermatozoa, suggesting that these genes participate in the spermatogenesis. Interestingly, the expression of zbtb family genes showed significantly changes in the RNA-seq data, and their expression patterns were further examined during spermatogenesis. The analysis of cellular localization of Eczbtb40 and the co-localization of Eczbtb40 and Eccyp17a1 in different gonadal stages suggested that Eczbtb40 might interact with Eccyp17a1 during spermatogenesis.
Conclusions
Our study, for the first time, investigated the transcriptome of the male germ cells from orange-spotted grouper, and identified functional genes, GO terms, and KEGG pathways involved in spermatogenesis. Furthermore, Eczbtb40 was first characterized and its role during spermatogenesis was predicted. These data will contribute to future studies on the molecular mechanism of spermatogenesis in teleosts.
Similar content being viewed by others
Background
In animals, spermatogenesis is a developmental process in which diploid male germ cells transform into haploid functional male gametes in a tight spatial and temporal organization. Spermatogonia develop into primary spermatocytes, and then primary spermatocytes are transformed into mature spermatozoa through two meiotic divisions. These processes are achieved by a complex interplay of genes and hormones [1]. During spermatogenesis, the testis contains different male germ cells and somatic cells. Revealing the transcriptome changes of these cells can facilitate our understanding of the mechanism of spermatogenesis in vertebrates. However, it is difficult to isolate the specific germ cells to investigate their transcriptome due to the complicated testis structure. Some approaches have already been used to quantify and localize unique genes in the testis during spermatogenesis, such as in situ hybridization [2, 3], immunohistochemistry, and serial analysis of gene expression [4, 5]. These methods can determine the spatial expression of specific gene during spermatogenesis, but cannot provide high throughput gene expression profiles in specific cell populations.
Laser capture microdissection (LCM) technology was developed in the late 1990s to obtaining targeted cell populations from tissue sections. This method was first used to study the gene expression in human cancer cells, and then widely used to study the spermatogenesis in rodents and fish species [6,7,8]. Although LCM can capture the targeted cells from the tissues, it is difficult to obtain enough RNA sample for traditional high-throughput RNA sequencing. In recent years, the single-cell RNA-sequencing technique has been developed, which only requires low amount of RNA to construct the transcriptome library. Therefore, high throughput gene expression analysis in specific germ cells during spermatogenesis can be performed by a combination of LCM and scRNA-seq.
Orange-spotted grouper is a protogynous hermaphroditic fish underlying sex change from female to male in its life history [9]. It has been considered as a good fish model for studying the sex reversal. As it is difficult to obtain the natural male fish, few studies on the spermatogenesis has been carried out in the orange-spotted grouper. In order to reveal the regulatory mechanism of spermatogenesis, in this study, LCM was applied to obtain the four germ cell types from the gonad of male orange-spotted grouper induced by MT implantation, including spermatogonia (SG), spermatocytes (SC), spermatids (ST), and spermatozoa (SZ), and the transcriptome sequencing was conducted subsequently. Through the transcriptomic analysis, zbtb (Zinc finger and BTB domain-containing protein) genes were found to be differentially expressed in different cell types. ZBTBs are an evolutionarily conserved family of transcription factors. Approximately 60 ZBTB proteins have been identified involving in diverse functions including development, differentiation, and oncogenesis [10,11,12]. In recent years, ZBTB16 has been found to play an essential role in spermatogenesis by controlling the self-renew and differentiation of spermatogonium [13,14,15,16]. Therefore, the zbtb family genes were further characterized and their expression was investigated during the process of spermatogenesis in orange-spotted grouper.
Results
Developmental stages of gonads during MT-induced spermatogenesis
Oocytes in the sham group remained in the primary-growth stage throughout the experimental period (Fig. 1a, b, d and f). In contrast, the fish in MT-implanted group underwent sex reversion from female to male. At the first week after MT implantation, the gonads were characterized by degeneration of oocytes and simultaneous proliferation of spermatogenic cysts (Fig. 1c). At the 2 weeks after MT implantation, the gonads entered into the intermediate transitional stage. Atretic oocytes were absorbed, and a mass of SC and ST were present (Fig. 1e). At the 3 weeks after MT implantation, the gonads entered into the late stage of spermatogenesis, and were occupied by SC and ST (Fig. 1g).
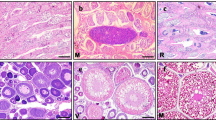
Gonadal histological morphology after MT treatment. a, b, d and f Histology of gonads in control fish. c, e and g Histology of gonads after MT implantation. PO, primary-growth stage oocyte; PVO, the cortical-alveolus stage oocyte; SG, spermatogonia; SC, spermatocyte; ST, spermatid; and SZ, spermatozoa. Scale bars = 50 um
Capture of the targeted cells
The morphology of SG, SC, ST, and SZ were characterized by hematoxylin and eosin (H&E, Fig. 2a) and quick staining (Fig. 2b). During spermatogenesis, the size of male germ cells decreases gradually, and the chromatin of the cells is condensed constantly [1]. Thus, different male germ cells can be distinguished by their morphology, size and density of chromatin [17]. In morphology, SG cell is irregular round, while SC, ST and SZ cells are more regular round. The size of four male cells become smaller gradually (SG > SC > ST > SZ). The staining of nuclei reflecting the density of chromatin is darker constantly in SG, SC, ST and SZ. Under the microscope, four different male germ cells were isolated by laser in PEN slices based on these characteristics. SG, SC and ST cells were captured from the gonad at the middle stage of spermatogenesis. SZ cells were obtained from the gonad at the late stage of spermatogenesis.
Gonadal structure by different staining methods. a Histological structure of gonad in MT treatment by H&E staining. b Histological structure of fresh gonad in MT treatment by quick staining. SG, spermatogonia; SC, spermatocyte; ST, spermatid; SZ, spermatozoa; and PO, primary-growth stage oocyte. Scale bars = 20 um
Validation of the sample quality
Five genes were used to validate the quality of the LCM-derived RNA samples (Fig. 3a). Ef1a is a reference gene and commonly used as an internal control for gene expression analysis. Its expression was detected in all four types of male germ cells at similar levels. Vasa is a germ cell marker [18] whose expression was also found in the four cell types. Slbp2 was specifically expressed in the oocyte of orange-spotted grouper [19], and no expression was detected in any male germ cells except for in the positive control (the gonad containing primary-growth stage oocytes). Dmrt1 was specifically expressed in the spermatogenic cells of orange-spotted grouper [20]. Here, its expression was only detected in SG and SC. Zbtb16 was expressed in spermatogonia specifically in orange-spotted grouper.
Expression of several sex-related genes in different tissues. a Expression of sex-related genes in four laser-captured cell types. The gene names were labeled on the left, and the length of genes was labeled on the right. M, marker 2000; C+, the cDNA of ovaries as a positive control; C-, template-free as a negative control; SG, spermatogonia; SC, spermatocyte; ST, spermatid; SZ, spermatozoa. b Tissue distribution of zbtb40 in eight tissues. Ef1a was used as the reference gene. M, marker 2000; 1, the whole brain; 2, heart; 3, head kidney; 4, liver; 5, kidney; 6, pituitary; 7, ovary; 8, testis
Quality of the transcriptomic data
Four cDNA libraries (SG, SC, ST, and SZ) were constructed. A total of 253,040, 890 raw reads were obtained, and a total of 244,984,338 clean reads were produced after removing low-quality reads and adapter sequences (Table 1). The average Q20 and Q30 values were 95.74 and 90.57%, respectively. The GC content was 44.08–47.47%.
Differentially expressed genes (DEGs) among the four cell types
Fragments per kilo base millions (FPKMs) were used to quantify the gene expression levels. The FPKM values of each gene in the four cell types were compared, respectively. There were 16,406 up-regulated genes and 11,054 down-regulated genes in SG compared to SC. A total of 15,845 up-regulated genes and 6895 down-regulated genes were identified in SC compared to ST. The ST had 8320 up-regulated genes and 15,797 down-regulated genes compared to SZ. Among the four cell types, 4483 DEGs were identified for further analysis.
GO and KEGG enrichment of DEGs
In the biological process and molecular function categories of the 4483 DEGs, cellular process (GO: 0009987) and binding (GO: 0005488) were the most enriched GO terms (Additional file 1: Figure S1). The top 20 pathways were listed from KEGG enrichment (Additional file 2: Table S1). Among them, apoptosis pathway, MAPK signaling pathway, and retinol metabolism pathway, were enriched during spermatogenesis (Fig. 4a).
The enrichment and expression analysis among the 4483 DEGs. a The top 20 pathways of KEGG analysis with the significant difference. b The heatmap based on the selected genes. Red indicated high expression and green low expression. The gene names were labeled on the right, and the validated genes by real-time PCR were marked by asterisk after the gene names. SG, spermatogonia; SC, spermatocyte; ST, spermatid; SZ, spermatozoa
Expression of functional genes putatively associated with sex differentiation and hormone metabolism
From the transcriptomic data, the expression of 20 genes putatively related to sex differentiation and hormone metabolism were analyzed (Fig. 5 and Additional file 1: Figure S2). Eight sex differentiation genes, sox11a (SRY-Box Transcription Factor 11a), nr0b1 (orphan nuclear receptor 0 B1), erα (estrogen reportor α), erβ (estrogen reportor β), wnt9 (Wingless-type MMTV integration site family member 9), gdf9 (oocyte-secreted growth differentiation factor 9), and bmp15 (bone morphogenetic protein 15), and thirteen hormone-related genes, cyp17a1 (steroid 17-alpha-hydroxylase/17, 20 lyase), hibadh (3-hydroxyisobutyrate dehydrogenase), dhrs11 (dehydrogenase/reductase family member 11), dhrs12 (dehydrogenase/reductase family member 12), p5cdh (delta-1-pyrroline-5-carboxylate dehydrogenase), cyp3a40 (cytochrome P450 3A40-like), and dhrs13 (dehydrogenase/reductase family member 13), which are involved in hormone metabolism were expressed in different cell types with FPKM values ranging from 2.62 to 604.7. Most of these genes were highly expressed in SG and SZ (Fig. 4b).
Validation of selected genes using real-time PCR during sex reversal. 0w, the female grouper before MT implantation; 1w, the early stage of sex reversal after 1week implantation; 2w, the middle stage of sex reversal after 2 weeks implantation; 3w, the late stage of sex reversal after 3 weeks MT implantation. The data was shown as mean ± SEM (n = 4 or 5) and the values with different asterisks were significantly different with a probability level <0.05 (P < 0.05). Ef1a was used as the reference gene. a nr0b1; b erα; c erβ; d wnt9; e gdf9; f bmp15; g cyp17a1; h zbtb40
Real-time PCR validation
Real-time PCR was performed to validate the expression patterns of 13 DEGs identified from the transcriptomic data. The expression of these genes was significantly changed during the spermatogenesis, which was largely consistent with their expression patterns in the transcriptomic data. Several female-related genes (erα, erβ, gdf9, and bmp15) increased significantly in the early stage of spermatogenesis (Fig. 5). The genes involved with hormone metabolism were also significantly increased during spermatogenesis (Additional file 1: Figure S2). These results indicate that the expressional analysis based on RNA-seq data was credible.
Molecular cloning and sequence analysis of zbtb40
Among the 4438 DEGs in four male germ cells, we found that several zbtb genes (zbtb1, zbtb22, zbtb40, and zbtb44) were differentially expressed. ZBTB family proteins are transcription factors participating in various important functions. Zbtb16 (plzf) was found to play an important role in the self-renewal and differentiation of the undifferentiated spermatogonia [13]. Whether the zbtb genes are involved in the regulation of spermatogenesis in orange-spotted grouper warrants further investigations. The expression of the zbtb genes (zbtb1, zbtb22, zbtb40, and zbtb44) was examined in spermatogenesis (Additional file 1: Figure S3). After further verification, the sequence and expression of zbtb40 were further characterized.
The open reading frame (ORF) of zbtb40 was cloned from the testis of orange-spotted grouper, and its sequence was submitted to GenBank (GenBank accession number, MN167853). The ORF of zbtb40 is 2400 bp in length, encoding a protein of 799 amid acids (Additional file 1: Figure S4). A phylogenetic tree showed that zbtb40 was clustered together with large yellow croaker (Larimichys crocea zbtb40) (Fig. 6a). The MEME web server was used to search the conserved motifs of zbtb40s. All zbtb40s were found to contain ten distinct conserved motifs (Fig. 6b). The DNA sequence of each motif site was displayed in Additional file 2: Table S2.
Phylogenetic relationships and conserved DNA motifs in zbtb40 genes from 14 species. a Phylogenetic tree of zbtb40s were conducted by MEGAX using the neighbor-joining likelihood method with 1000 bootstrap replicates. Numerals at nodes were bootstrap values. The species names are followed by sequence accession numbers. b The distribution of conserved motifs in ZBTB40s. The motifs are displayed in different colored boxes. The sequence information for each motif is provided in Additional file 2: Table S2
The expression pattern of zbtb40
The mRNA expression of zbtb40 in different tissues was examined by semi-quantitative PCR. The results revealed that zbtb40 is highly expressed in the testis, heart and pituitary, with low expression in the brain, head kidney, liver and ovary (Fig. 3b).
The expression pattern of zbtb40 during the MT-induced spermatogenesis was investigated by real-time PCR. The expression of zbtb40 was hardly detected in the gonad of sham group and 1 week after MT implantation. In the gonad of 2 weeks after MT implantation, the expression of zbtb40 was increased significantly compared to the sham group. However, the expression of zbtb40 was decreased in the 3 weeks after MT implantation (Fig. 5h).
In situ localization of zbtb40 in gonads during spermatogenesis
At the early stage of spermatogenesis, the zbtb40 mRNA signals were not detected in the oocytes but were observed in the male germ cells and somatic cells (Fig. 7b). At the middle stage of spermatogenesis, the zbtb40 mRNA is mainly expressed in SG and SC (Fig. 7c). At the late stage of spermatogenesis, zbtb40 was abundantly expressed in SG, SC, ST and SZ (Fig. 7d). In comparison, no zbtb40 signal was detected in the ovary of the sham group (Fig. 7a).
ISH analysis of zbtb40 mRNA expression in the gonads during the sex reversal. a Gonads without MT treatment, b gonads after 1 week of MT implantation, c gonads after 2 weeks of MT implantation, d gonads after 3 weeks of MT implantation. Red signals indicate zbtb40 and blue staining indicates nuclei. PO, primary-growth stage oocyte; ST, spermatids; SZ, spermatozoa. Scale bars = 50 μm
Co-localization of zbtb40 and cyp17a1 in ovary and testis
Given the similar expression patterns of zbtb40 and cyp17a1 during spermatogenesis, the cellular co-localization of two genes in the gonad was examined in ovary and testis by fluorescence in situ hybridization (FISH, Fig. 8). In ovary, cyp17a1 signals were clearly observed in the cytoplasm of oocytes where no zbtb40 signal was found (Fig. 8a-d). Cyp17a1 signals were located in the margin of oocyte, in the margin of nuclei in primary growth stage oocyte, and in the cytoplasm of cortical alveoli stage oocyte (Fig. 8b). In the gonad of the late stage of spermatogenesis, zbtb40 signals were located in the cytoplasm of SG and SC, which were overlapped with the cyp17a1 expression (Fig. 8e-h). Both zbtb40 and cyp17a1 had weaker expression in ST and SZ compared with SG and SC.
Co-localization of zbtb40 and cyp17a1 by FISH in the ovary (a-d) and testis induced by MT-implantation (e-h). (d) The merge image of (a-c). (h) The merge image of (e-g). i The magnified image of (h). Gonadal sections were stained red for zbtb40 mRNA, green for cyp17a1 mRNA, blue for DAPI, and orange for merge signal. PO, primary-growth stage oocyte; PVO, the cortical-alveolus stage oocyte; SG, spermatogonia; SC, spermatocyte; ST, spermatid. Scale bars = 25 μm
Discussion
In the present study, transcriptome analysis was used to profile the process of spermatogenesis based on the LCM technology. These data enhance our understanding on the mechanism of spermatogenesis in orange-spotted grouper as well as other vertebrates.
The whole process of LCM must produce sufficient amount of RNA with high quality to ensure the reliability of the transcriptome results. In this study, we modified a previous protocol [21] to handle slides and tissues, samples staining and capture the cells for preserving the RNA integrity throughout microdissection. To verify the quality of the four targeted cells, several cell markers were detected. The similar expression levels of ef1a in different cells indicated the homogeneity of concentration in four cell types. Vasa, a germ cell marker [22], is required for proper germ cells development. Its expression levels were decreased gradually with the progress of spermatogenesis, which is consistent with the expression pattern of vasa in the male germ cells of gibel carp [22] and brown-marbled grouper [18]. As an oocyte-specific marker [19, 23], slbp2 was used to distinguish the female germ cell and male germ cell. The non-expression of slbp2 in four cell types indicated that targeted cells didn’t contaminate by female germ cells. Dmrt1 was a spermatogenic cell marker [25]. Among numerous DEGs, it is interesting to note that some traditionally female related genes were highly expressed in the gonad after MT implantation, such as erα, erβ, gdf9 and bmp15. In mammals, estrogen receptors play crucial roles in spermatogenesis, e.g. the proliferation of gonocyte and spermatogonia, spermiation, sperm transport, and epididymal sperm maturation [26]. In eels, estrogen receptors are also expressed in the testis and are required for spermatogonial renewal [27, 28]. The high expression of estrogen receptors suggest that estrogen signals are also needed for spermatogenesis in grouper. GDF9 and BMP15, two members of the transforming growth factor beta family, were considered as female related genes promoting the oocyte maturation and folliculogenesis [29, 30]. However, several studies have reported the expression of gdf9 and bmp15 in testis, especially in male germ cells [DEGs analysis Prior to DEGs analysis, for each sequenced library, the read counts were adjusted by edgeR program package through one scaling normalized factor. Differential expression analysis of two conditions was performed using the DEGSeq R package (1.20.0). The P values were adjusted using the Benjamini & Hochberg method. Corrected P-value of 0.05 and log 2 (fold change) of 1 were set as the threshold for significant differential expression. GO terms with corrected P value less than 0.05 were considered to be significantly enriched by DEGs. The identified DEGs were conducted for enrichment analysis subsequently by GO: Termfinder software using the hypergeometric test [46, 47], and P-values were corrected using the Bonferroni method [48]. Significantly enriched GO terms were selected by Q value (Q < 0.05). KEGG is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information (http://www.genome.jp/kegg/). KOBAS software was used to test the statistical enrichment of differential expression genes in KEGG pathways (http://kobas.cbi.pku.edu.cn/m). To validate the RNA-seq data, the relative mRNA levels of 13 differentially expressed genes (nr0b1, erα, erβ, wnt9, gdf9, bmp15, cyp17a1, hibadh, dhrs11, dhrs12, p5cdh, cyp3a40, and dhrs13) were examined by quantitative real-time PCR in the gonad during spermatogenesis of orange-spotted grouper. Total RNA was extracted by TRIzol (Invitrogen, USA) and then 1 μg RNA from each sample was reverse transcribed with random primers by using the First Strand cDNA Synthesis Kit (Roche, USA) according to the manufacturer’s instruction. All mRNA quantification data were normalized to ef1a and presented as a relative control group. The specific primers used in this study were listed in Table 2.
Total RNA of the gonad was extracted by TRIzol (Invitrogen, USA). RNA was reversed to cDNA with First Strand cDNA Synthesis Kit (Roche, USA). Based on the cDNA fragments in RNA-seq data, specific upstream and downstream primers (Table 3) were designed. The primers were used to amplify the ORF of zbtb40. After PCR amplification, the band of the desired size was purified by the E.Z.N.A. Gel Extraction Kit (Omega, USA). The purified product was then subcloned into the pGEM-Easy vector (Fermentas, USA). According to the sequencing result, the ORF of zbtb40 was obtained.
The putative amino acid sequences were predicted by DNAMAN software. Nucleic acid phylogenetic analysis was conducted with MEGAX using the method of neighbor-joining method and the top ten motif sites were predicted by motif-based sequence analysis tools (MEME, http://meme-suite.org/). Total RNA from eight tissues (whole brain, heart, head kidney, liver, kidney, pituitary, ovary, and testis) was extracted. RNA was reversed to cDNA with First Strand cDNA Synthesis Kit (Roche, USA). The reverse transcription process was as follow: 37 °C for 15 min, 98 °C for 5 min, and 4 °C for 5 min. The PCR amplification regime was 35 cycles of 94 °C for 20 s, 55 °C for 10 s, and 72 °C for 20 s, followed by a further amplification at 72 °C for 5 min. The expression profile of zbtb40 in the gonad during spermatogenesis was detected by real-time PCR during MT-induced spermatogenesis. The methods were carried out as described above. The specific primers used in this study were listed in Table 3. The protocol of in situ localization (ISH) was referred to previous papers with minor modifications [49]. Probes of zbtb40 (628 bp) were synthesized by RNA DIG Labeling Kit (Roche, USA) according to the manufacturer’s instructions. After being permeabilized and acetylated, the cryosections were incubated by hybridization solution which contained 1 μg/ml probe, 20 x saline solution citrate (SSC) buffer, salmon sperm DNA, and deionized formamide, 50 x Denhart’s solution. After incubation for 12–16 h, the sections were washed by SSC and PBS buffer. The DIG label was tested with an alkaline phosphatase conjugated Flu-anti-DIG antibody (Roche Diagnostics; diluted 1:1000) and colored the signal with Fluorescence Systems (Roche, USA). Later the sections were counterstained by 4′ 6-diamidino-2-phenylindole (DAPI) for cell nuclear staining to confirm the number and status of germ cell. At last, sections were mounted with the Gold Anti-fade reagent (Invitrogen, USA) and imaged by laser scanning confocal microscope (Leica, TCS-SP5, Germany). The protocol of dual-label in situ hybridization was referred to previous study with minor modifications [50]. Briefly, the sample was hybridized by DIG-labeled zbtb40 RNA probes and biotin-labeled cyp17a1 RNA probes at the same time in 58 °C oven. After hybridization, the sections were washed by SSC and PBS buffer. The DIG label was tested with an alkaline phosphatase conjugated Flu-anti-DIG antibody, and the biotin-label was tested with an alkaline phosphatase conjugated streptavidin-POD antibody. The positive signals of ISH were developed by the TSA Plus Fluorescence Systems according to the product manual (Roche, USA). The signals of zbtb40 were red, and the signals of cyp17a1 were green. Cell nuclei were stained by DAPI (blue). Finally, the sections were mounted with the Gold Anti-fade reagent (Invitrogen, USA) and imaged with a microscope (Leica, TCS-SP5, Germany). All data were expressed as mean values ± SEM. Significant differences were checked by one-way analysis of variance (ANOVA) and student′s t-test was used, and a probability level less than 0.05 (P < 0.05) was used to indicate significance. All data were performed using GraphPad Prism5.0 (GraphPad Software, San Diego, CA) and analyzed by SPSS17.0 (SPSS, Chicago, IL, USA).GO and KEGG enrichment analysis of differentially expressed genes
Real-time PCR
Cloning and sequence analysis of zbtb40 cDNA
Tissue distribution of zbtb40 and its expression profile MT-induced spermatogenesis
In situ localization of zbtb40 in gonads during MT-induced spermatogenesis
Dual-label in situ hybridization of zbtb40 and cyp17a1 in gonads
Statistical analysis
Availability of data and materials
The datasets used and analyzed in the current study are available from the corresponding author on reasonable request. All relevant data are within the paper and the transcriptome data can be obtained from the Transcriptome Shotgun Assembly project DDBJ under accession number SSUB012433.
Abbreviations
- DEPC:
-
Diethyl pyrocarbonate
- DIG:
-
Digoxigenin
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes database
- PBS:
-
Phosphate buffered solution
- PEN:
-
Polyethylene naphthalate
References
Schulz RW, de Franca LR, Lareyre JJ, Le Gac F, Chiarini-Garcia H, Nobrega RH, Miura T. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165(3):390–411.
Eric L, Bittman LD, Huang L, Paroskie A. Period gene expression in mouse endocrine tissues. Am J Physiol Regul Integr Comp Physiol. 2003;285:R561–9.
Harvey S, Baudet ML, Murphy A, Luna M, Hull KL, Aramburo C. Testicular growth hormone (GH): GH expression in spermatogonia and primary spermatocytes. Gen Comp Endocrinol. 2004;139(2):158–67.
Chan WY, Lee TL, Wu SM, Ruszczyk L, Alba D, Baxendale V, Rennert OM. Transcriptome analyses of male germ cells with serial analysis of gene expression (SAGE). Mol Cell Endocrinol. 2006;250(1–2):8–19.
Yao J, Chiba T, Sakai J, Hirose K, Yamamoto M, Hada A, Kuramoto K, Higuchi K, Mori M. Mouse testis transcriptome revealed using serial analysis of gene expression. Mamm Genome. 2004;15(6):433–51.
Bonner RFE-BM, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Cell sampling: laser capture microdissection: molecular analysis of tissue. Science. 1997;278(21):1481–3.
Sluka P, O'Donnell L, Stanton PG. Stage-specific expression of genes associated with rat spermatogenesis: characterization by laser-capture microdissection and real-time polymerase chain reaction. Biol Reprod. 2002;67(3):820–8.
Sluka P, O'Donnell L, McLachlan RI, Stanton PG. Application of laser-capture microdissection to analysis of gene expression in the testis. Prog Histochem Cytochem. 2008;42(4):173–201.
Liu M, de Mitcheson YS. Gonad development during sexual differentiation in hatchery-produced orange-spotted grouper (Epinephelus coioides) and humpback grouper (Cromileptes altivelis) (Pisces: Serranidae, Epinephelinae). Aquaculture. 2009;287(1–2):191–202.
Beis D, Stainier DY. In vivo cell biology: following the zebrafish trend. Trends Cell Biol. 2006;16(2):105–12.
Stogios P, Downs G, Jauhal J, Nandra S, Privé G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82.
van Roy FM, McCrea PD. A role for Kaiso-p120ctn complexes in cancer? Nat Rev Cancer. 2005;5(12):956–64.
Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143(11):1893–906.
Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36(6):653–9.
Ozaki Y, Saito K, Shinya M, Kawasaki T, Sakai N. Evaluation of Sycp3, Plzf and Cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr Patterns. 2011;11(5–6):309–15.
Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–52.
Lacerda SM, Costa GM, de Franca LR. Biology and identity of fish spermatogonial stem cell. Gen Comp Endocrinol. 2014;207:56–65.
Boonanuntanasarn S, Bunlipatanon P, Ichida K, Yoohat K, Mengyu O, Detsathit S, Yazawa R, Yoshizaki G. Characterization of a vasa homolog in the brown-marbled grouper (Epinephelus fuscoguttatus) and its expression in gonad and germ cells during larval development. Fish Physiol Biochem. 2016;42(6):1621–36.
Wu X, Qu L, Li S, Guo Y, He J, Liu M, Liu X, Lin H. Molecular characterization and expression patterns of stem-loop binding protein (SLBP) genes in protogynous hermaphroditic grouper, Epinephelus coioides. Gene. 2019;700:120–30.
Wang Q, Liu Y, Peng C, Wang X, **ao L, Wang D, Chen J, Zhang H, Zhao H, Li S, et al. Molecular regulation of sex change induced by methyltestosterone -feeding and methyltestosterone -feeding withdrawal in the protogynous orange-spotted grouper. Biol Reprod. 2017;97(2):324–33.
Jensen E. Laser-capture microdissection. Antomical Rec (Hoboken, NJ: 2007). 2013;296(11):1683–7.
Holger Knaut FP, Bohmann K, Schwarz H, Nüsslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ Plasm and segregates asymmetrically before Germline specification. J Cell Biol. 2000;149(4):875–88.
He WWM, Liu Z, Li Z, Wang Y, Zhou J, Yu P, Zhang X, Li Z, Gui J. Oocyte-specific maternal Slbp2 is required for replication-dependent histone storage and early nuclear cleavage in zebrafish oogenesis and embryogenesis. RNA. 2018;24(12):1738–48.
**a W, Zhou L, Yao B, Li CJ, Gui JF. Differential and spermatogenic cell-specific expression of DMRT1 during sex reversal in protogynous hermaphroditic groupers. Mol Cell Endocrinol. 2007;263(1–2):156–72.
Li G-L, Qian H: Transcriptome using Illumina sequencing reveals the traits of spermatogenesis and develo** testes in Eriocheir sinensis. PLoS One 2017, 12(2):e0172478.
Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they Lead us? Endocr Rev. 1999;20(3):358–417.
Morini M, Penaranda DS, Vilchez MC, Tveiten H, Lafont AG, Dufour S, Perez L, Asturiano JF. The expression of nuclear and membrane estrogen receptors in the European eel throughout spermatogenesis. Comp Biochem Physiol A Mol Integr Physiol. 2017;203:91–9.
Takeshi Miura CM, Ohta T, Nader MR, Todo T, Y K. Estradiol-17β stimulates the renewal of spermatogonial stem cells in males. Biochem Biophys Res Commun. 1999;264:230–4.
Clelland E, Kohli G, Campbell RK, Sharma S, Shimasaki S, Peng C. Bone morphogenetic protein-15 in the zebrafish ovary: complementary deoxyribonucleic acid cloning, genomic organization, tissue distribution, and role in oocyte maturation. Endocrinology. 2006;147(1):201–9.
Sanfins A, Rodrigues P, Albertini DF. GDF-9 and BMP-15 direct the follicle symphony. J Assist Reprod Genet. 2018;35(10):1741–50.
He Z, Wu Y, **e J, Wang T, Zhang L, Zhang W. Growth differentiation factor 9 (Gdf9) was localized in the female as well as male germ cells in a protogynous hermaphroditic teleost fish, ricefield eel Monopterus albus. Gen Comp Endocrinol. 2012;178(2):355–62.
Zhao L, He J, Guo Q, Wen X, Zhang X, Dong C. Expression of growth differentiation factor 9 (GDF9) and its receptor in adult cat testis. Acta Histochem. 2011;113(8):771–6.
Yadav H, Lal B. BMP15 in catfish testis: cellular distribution, seasonal variation, and its role in steroidogenesis. Steroids. 2017;125:114–23.
Tang K, Wang L, ** Y, Yang W, Yang L. GDF9 affects the development and tight junction functions of immature bovine Sertoli cells. Reprod Domest Anim. 2017;52(4):640–8.
Beato MKJ. Steroid hormone receptors: an update. Hum Reprod Update. 2000;6(3):225–36.
Endo S, Miyagi N, Matsunaga T, Hara A, Ikari A. Human dehydrogenase/reductase (SDR family) member 11 is a novel type of 17beta-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun. 2016;472(1):231–6.
Sun C, Lu J, Yi G, Yuan J, Duan Z, Qu L, Xu G, Wang K, Yang N. Promising loci and genes for yolk and ovary weight in chickens revealed by a genome-wide association study. PLoS One. 2015;10(9):e0137145.
Kashiwada S, Kameshiro M, Tatsuta H, Sugaya Y, Kullman SW, Hinton DE, Goka K. Estrogenic modulation of CYP3A38, CYP3A40, and CYP19 in mature male medaka (Oryzias latipes). Comp Biochem Physiol C Toxicol Pharmacol. 2007;145(3):370–8.
Hinfray N, Nobrega RH, Caulier M, Baudiffier D, Maillot-Marechal E, Chadili E, Palluel O, Porcher JM, Schulz R, Brion F. Cyp17a1 and Cyp19a1 in the zebrafish testis are differentially affected by oestradiol. J Endocrinol. 2013;216(3):375–88.
Liu Y, Yao ZX, Bendavid C, Borgmeyer C, Han Z, Cavalli LR, Chan WY, Folmer J, Zirkin BR, Haddad BR, et al. Haploinsufficiency of cytochrome P450 17alpha-hydroxylase/17,20 lyase (CYP17) causes infertility in male mice. Mol Endocrinol. 2005;19(9):2380–9.
Zhai G, Shu T, **a Y, Lu Y, Shang G, ** X, He J, Nie P, Yin Z. Characterization of sexual trait development in cyp17a1-deficient Zebrafish. Endocrinology. 2018;159(10):3549–62.
Zhai G, Shu T, **a Y, ** X, He J, Yin Z. Androgen signaling regulates the transcription of anti-Müllerian hormone via synergy with SRY-related protein SOX9A. Sci Bull. 2017;62(3):197–203.
Shi Y, Zhang Y, Li S, Liu Q, Lu D, Liu M, Meng Z, Cheng CH, Liu X, Lin H. Molecular identification of the Kiss2/Kiss1ra system and its potential function during 17alpha-methyltestosterone-induced sex reversal in the orange-spotted grouper, Epinephelus coioides. Biol Reprod. 2010;83:63–74.
Blokhina O, Valerio C, Sokołowska K, Zhao L, Kärkönen A, Niittylä T, Fagerstedt K. Laser Capture Microdissection Protocol for Xylem Tissues of Woody Plants. Front Plant Sci. 2017;7:1965.
Golubeva Y, Salcedo R, Mueller C, Liotta LA, Espina V. Laser capture microdissection for protein and NanoString RNA analysis. Methods Mol Biol. 2013;931:213–57.
Flight RM, Wentzell PD. Potential bias in GO::TermFinder. Briefs Bioinformatics. 2009;10(3):289–94.
Boyle EI, Weng S, Gollub J, ** H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–5.
Aaltonen J, Laitinene M, Vuojolainen K, Jaatinen R, Horelli-Kuitunen N, Seppä L, Louhio H, Tuuri T, Sjöberg J, Bützow R, et al. Human growth differentiation factor 9 (GDF-9) and its novel homolog GDF-9B are expressed in oocytes during early Folliculogenesis. J Clin Endocrinol Metab. 1999;84(8):2744–50.
Xu H, Gui J, Hong Y. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev Dyn. 2005;233(3):872–82.
Guo Y, Wang Q, Li G, He M, Tang H, Zhang H, Yang X, Liu X, Lin H. Molecular mechanism of feedback regulation of 17beta-estradiol on two kiss genes in the protogynous orange-spotted grouper (Epinephelus coioides). Mol Reprod Dev. 2017;84(6):495–507.
Acknowledgments
Many thanks to Mr. Yu Chen, and this project was supported by Miss. Meifeng Liu and Ling Qu for their assistance in fish rearing and sample collection.
Funding
This work was supported by National Key Research and Development Program (2018YFD0900203), China Agriculture Research System (ARS-47), the Program of the China-ASEAN Maritime Cooperation Fund of the Chinese government (42000-41170002), Yang Fan Innovative & Entrepreneurial Research Team Project (No.201312H10), National Natural Science Foundation of China (No. 31672631), Natural Science Foundation of China (u1401213, 31802266), Special Fund for Agro-scientific Research in the Public Interest (201403011), and Huizhou Swan Project (2017021402310220). The funding body didn’t play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
XW and YY analyzed the sequencing data; XW, CYZ and YG collected and prepared the samples; XW wrote the manuscript; SSL and XCL supervised the study. All authors approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All the procedures in this manuscript had been approved by the Committee for Animal Experiments in Sun Yat-Sen University, China. The methods used in this study were carried out in accordance with the Laboratory Animal Management Principles of China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 : Figure S1.
GO classification analysis of all DEGs. Figure S2. Validation of selected genes using real-time PCR during sex reversal. Figure S3. Validation of zbtb genes using RT-PCR. Figure S4. The nucleotide sequences and deduced amino acid sequences of zbtb40.
Additional file 2 : Table S1.
The detailes of the top 20 KEGG enriched pathway. Table S2. The top 10 conserved sites among 14 species.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, X., Yang, Y., Zhong, C. et al. Transcriptome profiling of laser-captured germ cells and functional characterization of zbtb40 during 17alpha-methyltestosterone-induced spermatogenesis in orange-spotted grouper (Epinephelus coioides). BMC Genomics 21, 73 (2020). https://doi.org/10.1186/s12864-020-6477-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-020-6477-4