Abstract
Background
Dasypyrum villosum is an important wild species of wheat (Triticum aestivum L.) and harbors many desirable genes that can be used to improve various traits of wheat. Compared with other D. villosum accessions, D. villosum#4 still remains less studied. In particular, chromosomes of D. villosum#4 except 6V#4 have not been introduced into wheat by addition or substitution and translocation, which is an essential step to identify and apply the alien desired genes. RNA-seq technology can generate large amounts of transcriptome sequences and accelerate the development of chromosome-specific molecular markers and assisted selection of alien chromosome line.
Results
We obtained the transcriptome of D. villosum#4 via a high-throughput sequencing technique, and then developed 76 markers specific to each chromosome arm of D. villosum#4 based on the bioinformatic analysis of the transcriptome data. The D. villosum#4 sequences containing the specific DNA markers were expected to be involved in different genes, among which most had functions in metabolic processes. Consequently, we mapped these newly developed molecular markers to the homologous chromosome of barley and obtained the chromosome localization of these markers on barley genome. Then we analyzed the collinearity of these markers among D. villosum, wheat, and barley. In succession, we identified six types of T. aestivum-D. villosum#4 alien chromosome lines which had one or more than one D. villosum#4 chromosome in the cross and backcross BC3F5 populations between T. durum–D. villosum#4 amphidiploid TH3 and wheat cv. Wan7107 by employing the selected specific markers, some of which were further confirmed to be translocation or addition lines by genomic in situ hybridization (GISH).
Conclusion
Seventy-six PCR markers specific to chromosomes of D. villosum#4 based on transcriptome data were developed in the current study and their collinearity among D. villosum, wheat, and barley were carried out. Six types of Triticum aestivum-D. villosum#4 alien chromosome lines were identified by using 12 developed markers and some of which were further confirmed by GISH. These novel T. aestivum-D. villosum#4 chromosome lines have great potential to be used for the introduction of desirable genes from D. villosum#4 into wheat by chromosomal translocation to breed new wheat varieties.
Similar content being viewed by others
Background
Dasypyrum villosum (2n = 14, VV) is a cross-pollinating annual grass in the tribe Triticeae, which is native to the north-eastern part of the Mediterranean region (from southern France to the Caspian Sea), south-western Asia, Russia and the Caucasus areas. It is a vigorous ruderal plant that can grow in harsh and moisture-stressed environments [1]. D. villosum is resistant to many major wheat diseases including stripe rust (Puccinia striiformis), leaf rust (P. recondita), stem rust (P. graminis), powdery mildew (PM) caused by Blumeria graminis (Bgt), eyespot (Pseudocercosporella herpotrichoides) and wheat spindle streak mosaic virus transmitted by Polymyxa graminis [1,2,3,4,5]. In addition, D. villosum is tolerant to cold, salt, and drought stresses, has a high tillering rate, produces large amounts of seed, and has high protein content [1]. Therefore, D. villosum has great potential as a genetic source for improving cultivated wheat with more desirable traits. As of now, more than 300 D. villosum accessions have been collected [23].
D. villosum has many excellent genes, and the exact locations of some agronomic important genes on chromosomes are becoming clear (Table 1). Using the T. aestivum-D. villosum#2 translocation line T6V#2S·6AL, carrying Pm21, and T. aestivum-D. villosum#4 translocation line T6V#4S·6DL, carrying PmV, as PM resistance sources, some commercial wheat varieties including Neimai11, Neimai836, Yangmai18, Yangmai21, Yangmai97G59, Zhenmai9, Shimai14, Lantian27, **ghe9123 and Yangmai22 have been developed and released in different areas [24]. However, most studies on D. villosum have been focused on D. villosum#1, D. villosum#2, and D. villosum#3, and investigations on D. villosum#4 and D. villosum#5 have been relatively scarce.
Although SSR [25, 26], ISSR [27], EST-PCR [29] markers distributed on chromosomes 1V to 7V, especially on chromosome 6VS [24, 30,31,32,33,38] in D. villosum have been reported, most of these markers are developed based on the D. villosum#2 accession. Wide genetic diversity is known to exist among different original accessions such as D. villosum#2 and D. villosum#4. Previous studies have shown that there were structural divergences between D. villosum#2 and D. villosum#4 based on cytogenetic observations, 6VS-specific molecular markers, and differing reactions to wheat curl mite [24, 31, 39, 40]. The marker MBH1 can tag both Pm21 and PmV, from 6V#2S and 6V#4S, respectively, showing different sized amplification fragments [24]. In order to efficiently obtain addition, substitution, and translocation lines involved in each chromosome of T. aestivum-D. villosum#4, it is necessary to develop convenient molecular markers specific to each chromosome arm of D. villosum#4.
High-throughput RNA-seq technology can generate large amounts of transcriptome sequences and accelerate the development of chromosome-specific molecular markers. Recently, RNA-seq has been frequently used to develop molecular markers specific to chromosomes of wild relatives of cultivated wheat. For example, Thinopyrum intermedia genome-specific EST-SSR markers, Agropyron cristatum chromosome 6P-specific EST markers, Aegilops longissima chromosome-arm specific PCR markers and D. villosum#4 chromosome 6V#4S-specific PCR markers were developed using transcriptome data [31, 41,42,43]. Thereby, RNA-seq is a potential strategy to develop molecular markers specific to different chromosome arms of wild species of wheat.
In this study, we obtained the transcriptome data of D. villosum#4 by using RNA-seq and generated unigene sequences by transcriptome assembly after removing the transcripts from wheat by reference genome matching. Then, molecular primers specific to D. villosum#4 chromosomes 1V to 7V were designed based on the unigene sequences. Furthermore, six types of T. aestivum-D. villosum#4 candidate alien chromosome lines derived from the cross of T. durum–D. villosum#4 amphidiploid TH3 and wheat cv. Wan7107 followed by a backcross were identified by using a portion of the developed markers and genomic in situ hybridization (GISH). The results obtained in this study will be useful to breed addition, substitution and translocation lines of T. aestivum–D. villosum#4 and subsequent wheat varieties by marker assisted selection (MAS).
Results
Analysis of D. villosum#4 transcriptome data
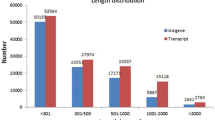
We carried out transcriptome sequencing of D. villosum#4 accession No. 1026, and obtained 7.14 G of clean bases and 47,619,528 clean reads in total. The rate of base mismatch was 0.02% (Table 2). Then, we spliced the clean reads and acquired a total of 74,517 transcript sequences and 47,382 unigene sequences (Fig. 1). The number of unigenes and transcripts sequences that had greater than 301 bp were 45,128 and 55,449, respectively. These sequences can be readily used for marker development. Seven databases were used for gene functional annotation of the 47,382 unigenes obtained in this study. The percentages of those unigenes annotated in Nr, Nt, Pfam, KOG/COG, Swiss-Prot, KEGG, and GO databases were 74.24, 4.31, 51.53, 28.5, 50.27, 25.19, and 52.29%, respectively. When comparing the unigene sequences to the non-redundant (Nr) library, we found that 29.8, 25.4, 17.3, 9.5, 8.6, and 9.5% of the unigene sequences were homologous to those in Hordeum vulgare, Aegilops tauschii, T. urartu, Brachypodium distachyon, T. aestivum, and other/unknown, respectively (Fig. 2).
Plant species classification of the transcriptome unigene matching to a non-redundant (Nr) library. 29.8% (red), 25.4% (green), 17.3% (navy blue), 9.5% (light blue), 8.6% (purple), and 9.5 (yellow) of the unigene sequences are homologous to those in Hordeum vulgare, Aegilops tauschii, T. urartu, Brachypodium distachyon, T. aestivum, and other/unknown, respectively
Primary selection of D. villosum#4 genome-specific markers
Those unigenes having the highest similarity to the sequences on the chromosomes of wheat groups 1 to 7 were considered candidate markers specific to D. villosum#4 chromosomes 1V to 7V. Primers were designed based on these unigene sequences, and further used to amplify the DNA of CS, one corresponding candidate line of the seven addition lines from DA1V#3 to DA7V#3, and D. villosum#4 accession No. 1026 for the screening of D. villosum#4 chromosome-specific amplicons. Figure 3 is an example of candidate markers specific to D. villosum#4 chromosome 5V. The amplification pattern of the candidate primers a, b, c, and d were verified at the next step (Fig. 3).
Amplification of candidate primers specific to D. villosum#4 targeted chromosomes designed by D. villosum#4 RNA-seq unigenes. M: marker 5000; 1: CS; 2: DA5V#3; 3: D. villosum#4 accession No. 1026. a: amplified by marker 5V-13; b: amplified by marker 5V-40F3R3; c: amplified by marker 5V-14; d: amplified by marker 5V-15
Confirmation of markers specific to D. villosum#4 chromosomes 1V to 7V
To develop specific molecular markers to each chromosome of D. villosum#4, seven T. aestivum-D. villosum#3 addition lines (DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3), No. 1026 and CS were amplified with the selected candidate primers, in which CS was used as the negative control, and No. 1026 was used as the positive control. Results showed that some primers amplified a unique band in D. villosum#4 and their corresponding addition line (one of the seven addition lines from DA1V#3 to DA7V#3) containing the targeted D. villosum#3 chromosome, and did not amplify the same band in CS (Fig. 4); some primers did not amplify any unique bands in CS and the seven addition lines (DA1V#3 to DA7V#3), only amplifying unique bands in D. villosum#4 No. 1026 (Additional file 1). There are abundant polymorphisms between the D. villosum#4 accession used for RNA-seq and the D. villosum#3 accession used as the alien chromosome donor of the seven addition lines in this study. However, some primers amplified unique alien band in an unexpected addition line, other than in the expected corresponding addition line. Finally, 76 D. villosum#4 chromosome-specific primers, most of which can specifically amplify bands in addition lines of DA1V#3 to DA7V#3, were obtained, including 1 in 1V#4, 9 in 2V#4, 27 in 3V#4, 10 in 4V#4, 15 in 5V#4, 13 in 6V#4, and 1 in 7V#4 (Additional file 2). These specific molecular markers can potentially be used to trace different chromosome arms of D. villosum#4 carrying useful genes for wheat breeding.
Amplification patterns of 12 different representative chromosome-specific primers in CS, 7 T. aestivum-D. villosum#3 addition lines, and D. villosum#4 accession No. 1026. M: marker 2000+; CS: Chinese Spring; 1: DA1V#3; 2: DA2V#3; 3: DA3V#3; 4: DA4V#3; 5: DA5V#3; 6: DA6V#3; 7: DA7V#3; 8: D. villosum#4 accession No.1026
Comparative analysis of the developed markers specific to D. villosum#4 genome with wheat and barley genome
As described above, we developed the markers specific to D. villosum#4 chromosomes 1V to 7V based on those unigenes which had the highest similarity to those sequences on the chromosomes of wheat groups 1 to 7. However, the D. villosum#4 transcriptome data (Fig. 2) showed that the D. villosum#4 unigene sequences had the most homology with Hordeum vulgare. To clarify the collinearity of the 76 newly developed specific PCR markers with the corresponding sequences among D. villosum, wheat, and barley. The chromosome localization of these markers on the barley was carried out by BLASTN (Additional file 2). It is illustrated that most corresponding sequences of the molecular markers specific to 1V to 6V have the same chromosome localization in wheat and barley, including one marker specific to 1V, 7 of 9 markers specific to 2V, 23 of 27 markers specific to 3V, 4 of 10 markers specific to 4V, 14 of 15 markers specific to 5V, 6 of 13 markers specific to 6V (Additional file 2).
Functional annotation of D. villosum#4 genes harboring the specific molecular markers
The 76 specific molecular markers were developed based on D. villosum#4 transcriptome sequences, and thus were expected to correspond to D. villosum#4 or D. villosum-associated genes. The sequences containing the 76 specific molecular markers were annotated in detail using the Nr, Nt, Pfam, KOG/COG, Swiss-prot, KEGG, and GO databases (Table 3). The annotations revealed that these markers were involved in different genes; most genes had functions in metabolic processes, and 22 genes had no annotation. For example, markers 6V-10, 6V-11, and 6V-12 were annotated to phyB activation tagged suppressor 1 (BAS1), which is a gene that regulates brassinosteroid levels and light responsiveness in Arabidopsis [44]. However, it is unknown whether a similar functioning gene may exist in D. villosum#4. Additionally, marker 5V-9 was annotated to E3 ubiquitin ligase which can catalyze the protein.
Application of D. villosum#4-specific markers in breeding new T. aestivum-D. villosum#4 alien chromosome lines
In order to develop new T. aestivum–D. villosum#4 alien chromosome lines which contain D. villosum#4 chromosomes 1V to 7V, a BC3F5 population consisting of 409 plants from the cross and backcross between the T. durum–D. villosum#4 amphidiploid TH3 and the wheat line Wan7107 was screened using 12 developed PCR markers specific to D. villosum#4 chromosomes 1V to 7V. Six types of T. aestivum-D. villosum#4 alien chromosome lines were obtained except 4V (Table 4). The specific primers 1V-1, 2V-1, 2V-5, 3V-4, 3V-23, 5V-3, 5V-15, 6V-2, 6V-10 and 7V-1 amplified different specific bands in D. villosum#4 No. 1026, the corresponding addition lines (DA1V#3, DA2V#3, DA3V#3, DA5V#3, DA6V#3, and DA7V#3), and the T. aestivum-D. villosum#4 candidate alien chromosome lines; however, they did not amplify any band in wheat line Wan7107 (Fig. 5).
Amplification patterns of 12 primers specific to D. villosum#4 1V to 7V. M: marker; 1–5: plants 201–1-1, 201–1-2, 201–1-3, 201–1-4, and 201–1-5, respectively; 6–9: plants 201–2-1, 201–2-3, 201–2-4, 201–2-5, respectively; 10, 23, and 36: W7107; 11, 24, and 37: DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3, respectively, from top to bottom in each column; 12, 25, and 38: D. villosum#4 accession No. 1026; 13–19: plants 203–1-1, 203–1-2, 203–1-3, 203–1-4, 203–1-5, 203–1-7, and 203–1-9, respectively; 20–22: plants 203–2-1, 203–2-2, and 203–2-3, respectively; 27–28: plants 204–2-2 and 204–2-3; 29–31: plants 204–3-1, 204–3-2, and 204–3-3, respectively; 32–35: plants 204–4-1, 204–4-2, 204–4-3, and 204–4-4, respectively
Due to the fact that markers 1V-1 and 5V-15 are located on D. villosum#4 chromosomes 1V and 5V, respectively, and markers 1V-1 and 5V-15 amplified specific bands for plants 201–1-1, 201–1-2, 201–1-3, 201–1-4, and 201–1-5, these plants most likely contain D. villosum#4 chromosomes 1V and 5V (Additional file 2). Then, the plant 201–1-3 were assayed by GISH using the genomic DNA of D. villosum#4 as a probe. As a result, two combined chromosomes between wheat and D. villosum#4 were clearly observed in this plant, indicating that plant 201–1-3 is a translocation line involved in D. villosum#4 1V and/or 5V (Fig. 6a).
Identification of the candidate T. aestivum–D. villosum#4 alien chromosome lines by GISH. Two translocated chromosomes between wheat and D. villosum#4 were clearly observed in plant 201–1-3 (a). A pair of translocated chromosomes between wheat and D. villosum#4 were clearly observed in plant 201–2-1 (b). One translocated chromosome between wheat and D. villosum#4 was clearly observed in plant 201–2-4 (c). Forty-three chromosomes including one D. villosum#4 chromosome were clearly observed in plant 204–2-3 (d). The scale bar in each photo is 10 μm in length
Marker 1V-1 amplified specific bands from plants 201–2-1, 201–2-4, and 201–2-5, indicating that these plants most likely contain D. villosum#4 chromosome 1V. Plants 201–2-1 and 201–2-4 were further assayed by GISH, and results indicated that two and one combined chromosomes between wheat and D. villosum#4 were clearly observed in the two plants, respectively, demonstrating that plants 201–2-1 and 201–2-4 are translocation lines of wheat chromosomes and D. villosum#4 1VL or 1VS (Fig. 6b and c). The marker and GISH detection results of these plants are consistent with their responses to exposure to Bgt. Plants 201–1-1, 201–1-2, 201–1-3, 201–1-4, and 201–1-5 showed high resistance to Bgt (Fig. 7). Based on our marker and GISH detection results, these plants most likely contain D. villosum#4 chromosome 5VL or 5VS. According to a previous study, a PM resistance gene Pm55 was identified to be located on D. villosum#2 chromosome 5VS [52]. Firstly, transcription data contain a large amount of transcript information, and the PCR marker based on transcription data is related to a definite trait and the corresponding transcript of this marker may be directly associated with the gene controlling this trait [53]. Secondly, the transcript sequences are highly conserved in their homologous genes, so the PCR markers developed based on transcription data are versatile in function and would be useful for modifying linkage maps or comparative maps among closely related species [54, 55].
The transfer of the foreign chromosomes into wheat through alien chromosome lines to confer desirable new traits also brings undesirable foreign genes. Nevertheless, wheat alien chromosome lines are very important foundational materials in chromosome engineering and wheat breeding programs. At present, research regarding the utilization of D. villosum focuses primarily on germplasm innovation. Wheat distant breeding using D. villosum will allow introduction of some desired genes into the general wheat background. This process is likely to be more successful when wheat-D. villosum alien chromosome lines are used as a bridge. Currently, wheat-D. villosum amphidiploid, addition, substitution, and translocation lines have been bred already. In order to efficiently identify D. villosum chromatin in the wheat background, including its targeted chromosomes and arms or fragments, it is necessary to develop a large number of markers specific to each chromosome arm or region of D. villosum. Current research on D. villosum, including marker development, is mostly focused on the accession D. villosum#2. Although some preliminary investigations on D. villosum#4 have been performed [31, 37, 39, 56], this genetic resource has not yet been fully exploited and utilized. Develo** markers specific to the D. villosum#4 genome will accelerate the application of the potential genes in D. villosum#4 and further expand the genetic background of wheat.
To develop molecular markers specific to D. villosum#4 using RNA-seq without knowing the chromosome locations of all the unigene sequences obtained by analyzing the transcriptome data, we initially submitted the unigene sequences to the URGI BLAST database https://urgi.versailles.inra.fr/blast/ to screen for unigene sequences which have high similarity to the sequences on the chromosomes of wheat groups 1 to 7. It was thus assumed that these unigene sequences were located on the chromosomes 1V to 7V of D. villosum#4. This might be a convenient approach to develop markers specific to the chromosomes of D. villosum#4 in the wheat background. Following this strategy, we located the D. villosum#4 unigenes to wheat genome, and then designed the specific molecular markers based on the differences between these unigenes in wheat and D. villosum#4 genomes.
Finally, we developed 76 PCR markers specific to the chromosome arms of D. villosum#4. Among them, enough markers specific to the chromosomes 2V, 3V, 4V, 5V, and 6V were obtained. The results revealed by transcriptome analysis indicated that these unigene sequences had the most homology with the corresponding sequences from Hordeum vulgare (Fig. 2). Further, we located the locations of the developed molecular markers on the barley H genome (Additional file 2), which will provide a reference for the localization of these molecular markers specific to D. villosum#4 in the wild relative species of common wheat. Based on the locations of these markers on the wheat groups 1–7 as compared with their locations on the barley H genome, we found that the locations of 55 markers specific to D. villosum#4 chromosomes were consistent with their chromosomal locations in wheat and barley (Additional file 2). These results suggest that there are some affinities among the three species. Eight markers including 2V-1, 2V-2, 4V-1, 4V-2, 4V-3, 6V-10, 6V-11, and 6V-12 display different chromosomal location in D. villosum#4, wheat, and barley, although these markers share the same chromosomal locations in wheat and barley (Additional file 2). Five markers of 4V-9, 4V-10, 5V-12, 6V-7, and 6V-9 have the same chromosomal location in D. villosum#4 and wheat, but different from that in barley. Four markers 3V-2, 3V-3, 3V-4, and 4V-4 have the same chromosomal location in D. villosum#4 and barley, but different from that in wheat (Additional file 2). Four markers 3V-1, 6V-4, 6V-13, and 7V-1 show different chromosomal location among the three plant species (Additional file 2). A similar phenomenon was also found in a previous report [29]. According to our results, the high homologous sequence of marker 2V-1 is located on chromosomes 5DS in wheat and 5H in barley, while it is not specifically amplified in the 5V addition line and it is specifically amplified in the 2V addition line on the contrary; the high homologous sequences of markers 3V-2, 3V-3, and 3V-4 are located on chromosome 5DL in wheat and 3H in barley, while they are not specifically amplified in the 5V addition line and they are specifically amplified in the 3V addition line; the high homologous sequences of markers 4V-1, 4V-2 and 4V-3 are located on chromosomes 7DS, 7AS and 5DL in wheat and 7H, 5H in barley, while they are not specifically amplified in the 7V, 5V addition line and they are specifically amplified in the 4V addition line (Additional file 2). This phenomenon suggested that there were some synteny among chromosomes 2V in D. villosum#4, wheat homoeologous groups 2, and 5, and barley 2H and 5H, between chromosomes 3V in D. villosum#4 and wheat homoeologous groups 3 and 5, and among chromosomes 4V in D. villosum#4, wheat homoeologous groups 4, 5 and 7, and barley 4H, 5H and 7H. Previously, it was reported that 59 4A ESTs could detect the loci on 5BL and 5DL, 72 4A ESTs could detect the loci on 7AS and 7DS, which confirmed the translocations involving chromosomes 4AL, 5BL, and 7AS in wheat [57]. This finding could explain part results of altered chromosomal location abovementioned. Taking together, the results aforementioned indicate that the collinearity of the developed markers on the chromosomal regions in D. villosum#4, wheat, and barley is interrupted. These structural differences reflect that the three plant species have undergone different evolutionary processes.
Update, more than 300 D. villosum accessions have been collected from different native habitats, and only a few (D. villosum #1 to D. villosum#5) have been used to develop wheat-D. villosum chromosome addition, substitution, and translocation lines [31, 39]. In this study, we were only able to develop one marker each for 1V and 7V. This phenomenon may be caused by the relationship between the chromosomes of D. villosum#4 and wheat, and the polymorphism between different sources of D. villosum. While develo** the markers specific to 1V and 7V of D. villosum#4, we found that most primers (86.67% in 1V and 70.59% in 7V) could not amplify any unique band in DA1V#3-DA7V#3, but that they could amplify bands in D. villosum#4. This indicated that there might be polymorphisms in the 1V and 7V between the accessions of D. villosum#4 and D. villosum#3. The specific reasons for this phenomenon need to be further studied. We will also try to map the correspondence D. villosum#4 unigenes to H genome for the development 1V- and 7V-specific markers in the near future.
Conclusions
Dasypyrum villosum is an important wild species of wheat and contain a lot of novel genes that can be used to improve agronomic traits of wheat. Most studies on D. villosum have focused on D. villosum#1, D. villosum#2, and D. villosum#3, whereas the number of investigations on D. villosum#4 is relatively less. We obtained the transcriptome data of D. villosum#4 by using RNA-seq. Then, 76 molecular markers specific to D. villosum#4 chromosomes 1V to 7V were developed based on transcriptome data. Furthermore, six types of T. aestivum-D. villosum#4 alien chromosome lines were identified by using 12 developed markers and some of which were further confirmed by GISH. These novel T. aestivum-D. villosum#4 chromosome lines have great potential to be used for the introduction of desirable genes from D. villosum#4 into wheat by chromosomal translocation to breed new wheat varieties.
Methods
Plant materials and cultivation
D. villosum#4 accession No. 1026 and wheat lines Chinese Spring (CS) and Wan7107, were maintained in our laboratory. T. aestivum-D. villosum#3 addition lines DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3 in CS background were kindly provided by Prof. Wenxuan Liu of Henan Agricultural University. A backcross BC3F5 population derived from the cross of T. durum–D. villosum#4 amphidiploid TH3 and wheat line Wan7107 was made by our laboratory. T. durum–D. villosum#4 amphidiploid TH3 (2n = 42, AABBVV) is derived from the cross between T. durum cv. Mexicali75 (2n = 28, AABB) and the D.villosum#4 accession No.1026 that originated from the former USSR. Wheat lines CS and Wan7107 and D. villosum#4 No. 1026, DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3 were grown in a greenhouse. The backcross population of plants from the cross of TH3 and Wan7107 were grown in the field.
RNA-seq and transcriptome assembly
Fresh leaves of D. villosum#4 accession No. 1026 were collected at seedling stage and RNA was extracted using TRIZOL (Thermo Fisher Scientific Inc., Shanghai, China). RNA quality, purity, integrity, and concentration were tested by the methods described in our previous publications [31, 43]. The RNA-seq was carried out at Bei**g Novogene Company, Bei**g, China (http://www.novogene.com/). Libraries for sequencing were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). Then, the libraries were sequenced and paired-end reads were generated on the Illumina Hiseq platform. Clean data were obtained by removing the low quality reads after the raw data was refined through in-house Perl scripts. The high quality clean data were used for further analyses. Transcriptome assembly was conducted using the Trinity software with the ‘--min_kmer_cov’ parameter for a mismatch rate set to 2 by default, and all other parameters were also set to the default [58].
Gene functional annotation
Gene function annotation was performed using Nr, Nt, Pfam, KOG/COG, Swiss-Prot, KO, and GO databases.
Development of markers specific to the genome of D. villosum#4 based on RNA-seq data
The strategy for screening candidate markers specific to the genome of D. villosum#4 was previously described by Li et al. [31]. Primers were designed using Primer Premier 6.0 (http://www.premierbiosoft.com/primerdesign/) based on those candidate unigene sequences which have the highest similarity to the sequences on the chromosomes of wheat groups 1 to 7.
DNA extraction and PCR amplification
Leaves were sampled from two-month-old seedlings of wheat lines CS and Wan7107, D. villosum accession#4 No. 1026, wheat-D. villosum#3 addition lines DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3, and the backcross population of TH3 and Wan7107, and stored at − 80 °C. Then, the genomic DNA for develo** specific makers and alien chromosome lines was isolated from all samples using a NuClean Plant Genomic DNA Kit (CW Bio Inc., China). And the genomic DNA used for probe in GISH was extracted from D. villosum#4 using the CTAB method. Finally, the DNA was dissolved in 50 μL ddH2O.
The PCR reactions of 15 μL volume included 100 ng of template DNA, 0.3 μL primer (10 μmol·L− 1), and 7.5 μL 2 × Taq MasterMix (containing Mg2+ and dNTP, CW Bio Inc., China). Amplification reaction was performed in a MiniAmp Plus thermal cycler (USA) with a PCR program including 95 °C for 5 min, 35 cycles of 20 s at 95 °C, 30 s at 60 °C, 1 min at 72 °C, and 8 min for 72 °C. Amplification products were run on a 2% agarose gel, and observed under UV light after Genecolour II (Gene-Bio, Bei**g, China) staining.
Chromosomes localization of D. villosum#4-specific markers on the barley H genome
The corresponding sequences of the 76 developed molecular markers specific to D. villosum#4 chromosomes were submitted onto website http://plants.ensembl.org/Multi/Tools/Blast?db=core to obtain chromosomal localization information through BLASTN searching.
GISH identification of new alien chromosome lines between T. aestivum and D. villosum#4
Wheat seeds were soaked in water containing 0.7% H2O2 for 24 h and put on three layers of moist filter paper in petri dishes. When the root tips grow to 2–3 cm in length, they were cut off and put into a 0.5 mL centrifuge tube (a hole was drilled on the lid of the tube before using). Then, the root tips were treated with nitrous oxide for 2 h in a special container and fixed in 90% acetic acid for 5 min. They were rinsed twice with sterile water. The apical milky parts of the root tips were excised and digested with cellulase and pectinase for 55 min. Cell spreading preparations at mitosis were made as previously described [43, 59, 60]. D. villosum#4 genomic DNA was labeled with fuorescein-12-dUTP as a probe using the nick translation method according to the manufacturer’s instructions. GISH was performed as previously description [43, 59,60,61]. Hybridization signals were captured under an Olympus BX-51 fluorescence microscope equipped with a charge-coupled device system.
Abbreviations
- Bgt :
-
Blumeria graminis f. sp. tritici
- COG:
-
Clusters of orthologous groups of proteins
- CS:
-
Chinese Spring
- DA:
-
Disomic addition
- EST:
-
Expressed sequence tag
- GISH:
-
Genomic in situ hybridization
- GO:
-
Gene ontology
- HMW:
-
High molecular weight
- ISSR:
-
Inter-simple sequence repeats
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KOG:
-
Eukaryotic ortholog groups
- LMW:
-
Low molecular weight
- MAS:
-
Marker assisted selection
- Nr:
-
NCBI non-redundant protein sequences
- Nt:
-
NCBI nucleotide sequences
- Pfam:
-
Protein family
- PM:
-
Powdery mildew
- SNPs:
-
Single-nucleotide polymorphisms
- SSR:
-
Simple sequence repeats
- STS:
-
Sequence-tagged site
References
Agnieszka G. The genus Dasypyrum—part 2. Dasypyrum villosum—a wild species used in wheat improvement. Euphytica. 2006;152(3):441–54.
Minelli S, Ceccarelli M, Mariani M, De Pace C, Cionini PG. Cytogenetics of Triticum·Dasypyrum hybrids and derived lines. Cytogenet Genome Res. 2005;109(1–3):385–92.
Yildirim A, Jones SS, Murray TD, Line RF. Evaluation of Dasypyrum villosum populations for resistance to cereal eyespot and stripe rust pathogens. Plant Dis. 2000;84(1):40–4.
Zhang QP, Li Q, Wang XE, Wang HY, Lang SP, Wang YN, et al. Development and characterization of a Triticum aestivum–Haynaldia villosa translocation line T4VS·4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica. 2005;145(3):317–20.
Chen Q, Conner RL, Li H, Laroche A, Graf RJ, Kuzyk AD. Expression of resistance to strip rust, powdery mildew and the wheat curl mite in Triticum aestivum×Haynaldia villosa lines. Can J Plant Sci. 2002;82(2):451–6.
Zhang RQ, Fan YL, Kong LN, Wang ZJ, Wu JZ, **ng LP, et al. Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor Appl Genet. 2018;131:2613–20.
Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, et al. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Gene. 2011;123(1):159–67.
De Pace C, Vaccino P, Cionini G, Pasquini M, Bizzarri M, Qualset CO. Dasypyrum. In: Kole C, editor. Wild crop relatives: genomic and breeding resources, cereals, Vol 1, chapter 4. Heidelberg: Springer; 2011. p. 185–292.
Liu C, Qi LL, Liu WX, Zhao WC, Wilson J, Friebe B, et al. Development of a set of compensating Triticum aestivum–Dasypyrum villosum Robertsonian translocation lines. Genome. 2011;54:836–44.
Zhang RQ, Zhang MY, Wang XE, Chen PD. Introduction of chromosome segment carrying the seed storage protein genes from chromosome 1V of Dasypyrum villosum showed positive effect on bread-making quality of common wheat. Theor Appl Genet. 2014;127(3):523–33.
Li H, Chen X, **n ZY, Ma YZ, Xu HJ, Chen XY, et al. Development and identification of wheat–Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed. 2005;124(2):203–5.
Sears ER. Addition of the genome of Haynaldia villosa to Triticum avestivum. Am J Bot. 1953;40(3):168–74.
Hyde BB. Addition of individual Haynaldia villosa chromosomes to hexaploid wheat. A J Bot. 1953;40(3):174–82.
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet. 1995;91(6–7):1125–8.
Liu DJ, Chen PD, Pei GZ, Wang YN, Qiu BX, Wang SL. Transfer of Haynaldia villosa chromosomes into Triticum aestivum. In: Miller TE, RMD K, editors. Proceedings of the 7th international wheat genetic symposium. Cambridge, UK: the 7th international wheat genetic symposium; 1988;1:355–61.
Qi LL, Chen PD, Liu DJ, Zhou B, Zhang SZ. Development of translocation lines of Triticum aestivum with powdery mildew resistance introduced from Haynaldia villosa. In: Proc of the 8th intern wheat genet symp. Be**g: Chinese Agricultural Scientech Press; 1995. p. 333–7.
Zhang RQ, Cao YP, Wang XE, Feng YG, Chen PD. Development and characterization of a Triticum aestivum–D. villosum T5VS.5DL translocation line with soft grain texture. J Cereal Sci. 2010;51(2):220–5.
Zhang RQ, Hou F, Feng YG, Zhang W, Zhang M, Chen PD. Characterization of a Triticum aestivum–Dasypyrum villosum T2VS·2DL translocation line expressing a longer spike and more kernels traits. Theor Appl Genet. 2015;128(12):2415–25.
Zhang RQ, Sun BX, Chen J, Cao AZ, **ng LP, Feng YG, et al. Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor Appl Genet. 2016;129(10):1975–84.
Zhao WC, Qi LL, Gao X, Zhang GS, Dong J, Chen QJ, et al. Development and characterization of two new Triticum aestivum-Dasypyrum villosum Robertsonian translocation lines T1DS•1V#3L and T1DL•1V#3S and their effect on grain quality. Euphytica. 2010;175(3):343–50.
Dong J, Yang H, Zhao WC, Li XY, Chen QJ, Gao X. Agronomic traits and grain quality of Chinese spring-Dasypyrum villosum translocation lines T1DL·1VS and T1DS·1VL. Acta Agron Sin. 2013;39(8):1386–90.
Chen X, Shi AN, Shang LM, Steven L, Paul MJ. The resistance reaction of Haynaldia villosa to powdery mildew isolates and its expression in wheat background. Acta Phytopath Sin. 1997;27(1):17–22.
Cho S, Garvin DF, Muehlbauer GJ. Transcriptome analysis and physical map** of barley genes in wheat–barley chromosome addition lines. Genetics. 2006;172(2):1277–85.
Bie TD, Zhao RH, Zhu SY, Chen SL, He HG. Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol Breeding. 2015;35:189.
Liu SB, Tang ZH, You MS, Li BY, Song JM, Liu GT. Characterization of chromosome 1V specific SSR molecular markers from Haynaldia villosa. Acta Agron Sin. 2004;30(2):138–42.
Zhang W, Gao AL, Zhou B, Chen PD. Screening and applying wheat microsatellite markers to trace individual Haynaldia villosa chromosomes. Acta Genet Sin. 2006;33(3):236–43.
Tang ZQ, Yang ZJ, Li GR, Liu C, Liu ZH, Ren ZL. Development of ISSR marker specific for Dasypyrum chromosome 5V and detection of its related species. J Agric Biotechnol. 2007;15(5):799–804.
Zhao RH, Wang HY, Jia Q, **ao J, Yuan CX, Zhang YJ, et al. Development of EST-PCR markers for the chromosome 4V of Haynaldia villosa and their application in identification of 4V chromosome structural Aberrants. J Intensiv Agric. 2014;13(2):282–9.
Zhang RQ, Yao RN, Sun DF, Sun BX, Feng YG, Zhang W, et al. Development of V chromosome alterations and physical map** of molecular markers specific to Dasypyrum villosum. Mol Breed. 2017;37(5):1–11.
Blanco A, Resta P, Simeone R, Parmar S, Shewry PR, Sabelli P, et al. Chromosomal location of seed storage protein genes in the genome of Dasypyrum villosum (L.) Candargy. Theor Appl Genet. 1991;82:358–62.
Li SJ, Lin ZS, Liu C, Wang K, Du LP, Ye XG. Development and comparative genomic map** of Dasypyrum villosum 6V#4S-specific PCR markers using transcriptome data. Theor Appl Genet. 2017;130(10):2057–68.
Chen YP, Wang HZ, Cao AZ, Wang CM, Chen PD. Cloning of a resistance gene analog from wheat and development of a codominant PCR marker for Pm21. J Integr Plant Biol. 2006;48(6):715–21.
Wang CM, Bie TD, Chen QZ, Cao AZ, Chen PD. Development and application of molecular markers specific to chromosome 6VS of Haynaldia villosa (in Chinese). Acta Agron Sin. 2007;33(10):1595–600.
Song W, **e CJ, Du JK, **e H, Liu Q, Ni ZF, et al. A “one-marker-for-two-genes” approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat. Mol Breed. 2009;23(3):357–63.
He HG, Zhu SY, Sun WH, Gao DR, Bie TD. Efficient development of Haynaldia villosa chromosome 6VS-specific DNA markers using a CISP-IS strategy. Plant Breed. 2013;132(3):290–4.
Chen PD, You CF, Hu Y, Chen SW, Zhou B, Cao AZ, et al. Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol Breed. 2013;31(2):477–84.
Zhang YL, Wang MJ, Zhang Y, Chu CP, Lin ZS, Xu QF, et al. Development and application of functional markers specific to powdery mildew resistance on chromosome arm 6VS from different origins of Haynaldia villosa (in Chinese). Acta Agron Sin. 2012;38(10):1827–32.
He HG, Zhu SY, Jiang ZN, Ji YY, Wang F, Zhao RH, et al. Comparative map** of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor Appl Genet. 2016;129(4):819–29.
Liu C, Ye XG, Wang MJ, Li SJ, Lin ZS. Genetic behavior of Triticum aestivum–Dasypyrum villosum translocation chromosomes T6V#4S·6DL and T6V#2S·6AL carrying powdery mildew resistance. J Integr Agr. 2017;16(10):2136–44.
Li HJ, Conner RL, Chen Q, Jia X, Li H, Graf RJ, et al. Different reactions to the wheat curl mite and wheat streak mosaic virus in various wheat–Haynaldia villosa 6V and 6VS lines. Plant Dis. 2002;86(4):423–8.
Cui Y, Bao YG, Wang HG, Li XF. Development of specific molecular markers for Thinopyrum intermedium using RNA-seq data. J Triticeae Crop. 2016;36(6):699–707.
Dai C, Zhang JP, Wu XY, Yang XM, Li XQ, Liu WH, et al. Development of EST markers specific to Agropyron cristatum chromosome 6P in common wheat background. Acta Agron Sin. 2012;38(10):1791–801.
Wang KY, Lin ZS, Wang L, Wang K, Shi QH, Du LP, et al. Development of a set of PCR markers specific to Aegilops longissima chromosome arms and application in breeding a translocation line. Theor Appl Genet. 2018;131(1):13–25.
Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96(26):15316–23.
Alagna F, D’Agostino N, Torchia L, Servili M, Rao R, Pietrella M, et al. Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. BMC Genomics. 2009;10:399.
Novaes E, Drost DR, Farmerie WG, Pappas GJ, Grattapaglia D, Sederoff RR, et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9(1):312.
Wang ET, Sandberg R, Luo SJ, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6.
Li AL, Zhang RZ, Pan L, Tang LC, Zhao GY, Zhu MZ, et al. Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PLoS One. 2011;6(12):e28810.
Jia CL, Zhang Y, Zhu L, Zhang R. Application progress of transcriptome sequencing technology in biological sequencing. Mol Plant Breed. 2015;10:2388–94.
Zhang JC, Zheng HY, Li YW, Li HJ, Liu X, Qin HJ, et al. Coexpression network analysis of the genes regulated by two types of resistance responses to powdery mildew in wheat. Sci Rep. 2016;6:23805.
Franssen SU, Shrestha RP, Brautigam A, Bornberg-Bauer E, Weber AP. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics. 2011;12(1):227.
Li XB, Cui HR, Zhang ML. Molecular markers derived from EST: their development and applications in comparative genomics. Biodivers Sci. 2006;14(6):541–7.
Bozhko M, Riegel R, Schubert R, Muller-Starck G. A cyclophilin gene marker confirming geographical differentiation of Norway spruce populations and indicating viability response on excess soil-born salinity. Mol Ecol. 2003;12(11):3147–55.
Lan TH, DelMonte TA, Reischmann KP, Hyman J, Kowalski SP, McFerson J, et al. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 2000;10(6):776–88.
Varshney RK, Sigmund R, Borner A, Korzun V, Stein N, Sorrells ME, et al. Interspecifc transferability and comparative map** of barley EST-SSR markers in wheat, rye and rice. Plant Sci. 2005;168(1):195–202.
Lin ZS, Zhang YL, Wang K, Li JR, Xu QF, Chen X, et al. Isolation and molecular analysis of genes Stpk-V2 and Stpk-V3 homologous to powdery mildew resistance gene Stpk-V in a Dasypyrum villosum accession and its derivatives. J Appl Genet. 2013;54(4):417–26.
Miftahudin RK, Ma XF, Mahmoud AA, Layton J, Rodriguez Milla MA, Chikmawati T, et al. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics. 2004;168(2):651–63.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52.
Han FP, Liu B, Fedak G, Liu Z. Chromosomal variation, constitution of five partial amphiploids of wheat–Thinopyrum intermedium detected by GISH, seed storage protein marker and multicolor GISH. Theor Appl Genet. 2004;109(5):1070–6.
Yuan J, Guo X, Hu J, Lv ZL, Han FP. Characterization of two CENH3 genes and their roles in wheat evolution. New Phytol. 2015;206(2):839–51.
Guo X, Su HD, Shi QH, Fu SL, Wang J, Zhang XQ, et al. De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PLoS Genet. 2016;12(4):e1005997.
De Pace C, Snidaro D, Ciaffi M, Vittori D, Ciofo A, Cenci A, et al. Introgression of Dasypyrum villosum chromatin into common wheat improves grain protein quality. Euphytica. 2001;117(1):67–75.
Shewry PR, Parmar S, Pappin DJC. Characterization and genetic control of the prolamins of Haynaldia villosa: relationship to cultivated species of the Triticeae (rye, wheat and barley). Biochem Genet. 1987;25:309–25.
Liu CJ, Chao S, Gale MD. Wsp-1, a set of genes controlling water-soluble proteins in wheat and related species. Genet Res. 1989;54(3):173–81.
Zhao CZ, Lv XD, Li YH, Li F, Geng MM, Mi YY, et al. Haynaldia villosa NAM-V1 is linked with the powdery mildew resistance gene Pm21 and contributes to increasing grain protein content in wheat. BMC Genet. 2016;17:82.
He HG, Zhu SY, Zhao RH, Jiang ZN, Ji YY, Ji J, et al. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant. 2018;11:879–82.
**ng LP, Hu P, Liu JQ, Witek K, Zhou S, Xu JF, et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR that confers powdery mildew resistance in wheat. Mol Plant. 2018;11(6):874–8.
Cao AZ, **ng LP, Wang XY, Yang XM, Wang W, Sun YL, et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci U S A. 2011;108(19):7727–32.
Zhu YF, Li YB, Fei F, Wang ZK, Wang W, Cao AZ, et al. E3 ubiquitin ligase gene CMPG1–V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J. 2015;84(1):154–68.
Faheem M, Li YB, Arshad M, Cheng JY, Zhao J, Wang ZK, et al. A disulphide isomerase gene (PDI-V) from Haynaldia villosa contributes to powdery mildew resistance in common wheat. Sci Rep. 2016;6:24227.
Wang ZK, Cheng JY, Fan AQ, Zhao J, Yu ZY, Li YB, et al. LecRK-V, an L-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol J. 2018;16(1):50–62.
Yildirim A, Jones SS, Murray TD. Map** a gene conferring resistance to Pseudocercosporella herpotrichoides on chromosome 4V of Dasypyrum villosum in a wheat background. Genome. 1998;41:1–6.
Zhao RH, Wang HY, ** of the wheat yellow mosaic virus resistance gene from Haynaldia villosa. Theor Appl Genet. 2013;126(12):2921–30.
Zhang RQ, Feng YG, Li HF, Yuan HX, Dai JL, Cao AZ, et al. Cereal cyst nematode resistance gene CreV effective against Heterodera filipjevi transferred from chromosome 6VL of Dasypyrum villosum to bread wheat. Mol Breeding. 2016;36:122.
Zhang RQ, Feng YG, Hou F, Chen SL, Bie TD, Chen PD. The genetic effect of wheat-H. villosa T5VS.5DL translocated chromosome on agronomic characteristics, quality, and powdery mildew resistance of common wheat. Scientia Agricultura Sin. 2015;48(6):1041–51.
Acknowledgments
We thank Prof. Wenxuan Liu at Henan Agricultural University, China, for providing a set of T. aestivum-D. villosum#3 disomic chromosome addition lines (DA1V#3, DA2V#3, DA3V#3, DA4V#3, DA5V#3, DA6V#3, and DA7V#3), and Prof. Fangpu Han at Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China, for providing the experimental facilities for GISH analysis of the developed T. aestivum-D. villosum#4 alien chromosome lines.
Funding
This research was financially supported by the National Key Research and Development Program of China (2016YFD0102001 and 2016YFD0102002), the National Natural Science Foundation of China (31771788), and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences. The funders had no role in the designing and conducting of this study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Author information
Authors and Affiliations
Contributions
The experiment was conceived by XGY and ZSL. SJL and ZSL analyzed the transcriptome data. SJL, JW, KYW, and KW developed the molecular markers. ZSL, JNC, and XGY made the crosses and backcrosses between T. durum–D. villosum#4 amphidiploid TH3 and wheat cv. Wan7107. SJL and JNC screened the backcrossing population using molecular markers for alien chromosome lines. JW identified the selected T. aestivum-D. villosum#4 alien chromosome lines by GISH. ZSL, ZFN, and XGY tested the T. aestivum-D. villosum#4 alien chromosome lines for PM resistance. LPD managed the materials in greenhouse and field and helped in DNA extraction. The manuscript was drafted by SJL, XGY, and ZSL, and corrected and approved by all authors.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Amplification patterns of six 1V and 7V candidate chromosome-specific primers in Chinese (CS), seven T. aestivum-D. villosum#3 addition lines, and D. villosum#4 accession No. 1026. M: marker 2000+; CS: Chinese Spring; 1: DA1V#3; 2: DA2V#3; 3: DA3V#3; 4: DA4V#3; 5: DA5V#3; 6: DA6V#3; 7: DA7V#3; 8: D. villosum#4 accession No.1026. (DOCX 95 kb)
Additional file 2:
Localization of the 76 developed PCR markers in wheat, barley, and D. villosum#4, and their primer sequences and amplified sizes. 1: Chromosomal localization of the corresponding homologous sequences of the developed markers in wheat; 2: Chromosomal localization of the corresponding homologous sequences of the developed markers in barley; 3: Chromosomal localization of the corresponding sequences of the developed markers in D. villosum#4. (DOCX 24 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, S., Wang, J., Wang, K. et al. Development of PCR markers specific to Dasypyrum villosum genome based on transcriptome data and their application in breeding Triticum aestivum-D. villosum#4 alien chromosome lines. BMC Genomics 20, 289 (2019). https://doi.org/10.1186/s12864-019-5630-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5630-4