Abstract
Background
Modern genome sequencing leads to an ever-growing collection of genomic annotations. Combining these elements with a set of input regions (e.g. genes) would yield new insights in genomic associations, such as those involved in gene regulation. The required data are scattered across different databases making a manual approach tiresome, unpractical, and prone to error. Semi-automatic approaches require programming skills in data parsing, processing, overlap calculation, and visualization, which most biomedical researchers lack. Our aim was to develop an automated tool providing all necessary algorithms, benefiting both bioinformaticians and researchers without bioinformatic training.
Results
We developed overlap** annotated genomic regions (OGRE) as a comprehensive tool to associate and visualize input regions with genomic annotations. It does so by parsing regions of interest, mining publicly available annotations, and calculating possible overlaps between them. The user can thus identify location, type, and number of associated regulatory elements. Results are presented as easy to understand visualizations and result tables. We applied OGRE to recent studies and could show high reproducibility and potential new insights. To demonstrate OGRE’s performance in terms of running time and output, we have conducted a benchmark and compared its features with similar tools.
Conclusions
OGRE’s functions and built-in annotations can be applied as a downstream overlap association step, which is compatible with most genomic sequencing outputs, and can thus enrich pre-existing analyses pipelines. Compared to similar tools, OGRE shows competitive performance, offers additional features, and has been successfully applied to two recent studies. Overall, OGRE addresses the lack of tools for automatic analysis, local genomic overlap calculation, and visualization by providing an easy to use, end-to-end solution for both biologists and computational scientists.
Similar content being viewed by others
Background
Modern genome sequencing produces ever-growing numbers of large genomic datasets for multiple organisms. Consequently, processing and putting these data into biologically meaningful context remains a challenge. Databases like Ensembl and UCSC store this information and list an increasing amount of annotation data [1, 2]. In the human genome, there are now over 20,000 known protein-coding, 40,000 micro (miRNA), and 19,000 long non-coding (lncRNA) RNA genes, 252,000 transcripts, 30 million CpG sites, and 5 million single nucleotide polymorphisms (SNPs) [3, 4]. Apart from this, the amount of available data on the location of epigenetic markers—such CpG islands (CGI), histone modifications, and chromatin 3D structure—and regulatory regions—e.g. promoters and enhancers—has also grown in recent years [5, 6]. These genomic elements can occupy regions from a few base pairs up to several mega base pairs and are not randomly distributed throughout the genome. Regions that are overlap** or neighboring each other might interplay and perform potential regulatory functions. A prime example are promoter regions, typically located upstream from the transcription start site (TSS) [7] and involved in the regulation of gene expression (8, 9), which often include transcription factor binding sites (TFBS) and CGIs.
Techniques such as RNA sequencing, methylome analysis, chromatin immunoprecipitation (ChIP) sequencing, and whole genome association studies often result in a set of candidate genes or a collection of interesting genomic regions, which need to be further investigated by researchers who are not always trained in using bioinformatic and data processing tools. Especially, the validation and further downstream analysis of candidate genes resulting from differential gene expression analysis benefits from information about the various regulatory elements controlling gene transcription. This requires acquiring, mining, and parsing multiple datasets for the overlap of regions with e.g. promoters, TFBS, CGIs, and other regulatory elements. A manual approach to identify TFBS and CGIs within a gene’s promoter region, for instance, is not trivial. The first step—to mine published annotations for overlaps with a list of candidate genes—requires obtaining genomic coordinates for each candidate and manually searching for the features of interest in one of the various genome browsers (e.g. UCSC or Ensembl). One would then need to make sure to obtain annotations for all regulatory elements of interest and visualize them using appropriate software (e.g. IGV browser), which might not be easily achievable. In the next step, overlap** or neighboring regulatory elements need to be identified and annotated. Finally, genomic locations, results, and graphics must be ponderously exported for further processing. This manual approach can be tiresome, unreliable, and prone to errors, especially for long candidate lists. It further leads to non-comparability and varying results depending on the person conducting the manual analysis. In addition, most researchers might not have the bioinformatics expertise necessary for retrieving and visualizing the results. Nevertheless, this kind of analysis remains a key element in understanding the interplay between regulatory elements and thus the observed gene expression changes under different experimental conditions. Therefore, a tool allowing researchers to identify the presence of regulatory elements for a set of genomic regions in a user friendly and platform-independent way is urgently needed. To our knowledge, no software tool is available that allows the automation of these tasks, and such a complex analysis still requires the help of a bioinformatician or computational scientist to do the necessary data parsing and programming. Therefore, we developed OGRE (Overlap** annotated Genomic Regions) as a user friendly and easily accessible tool to perform automatic overlap analysis, export tabular results, and visualize genomic regions based on publicly available annotations. In addition, the user interface SHREC (SHiny interface for REgion Comparison) provides accessibility for biologists without computational training.
Implementation
Workflow
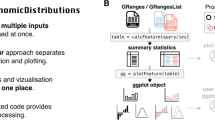
Internally OGRE methods are structured in three modules listed as follows: (1) Dataset module, (2) Processing module, and (3) Visualization module (Fig. 1). We further define an OGREDataSet as a list of datasets with additional metadata information that serves as input for each module. The Dataset module reads user-generated local tabular data like .CSV and .GFF files which often result from OMICS experiments. Once the user defines a directory, it is scanned for suitable file types, which are attached to the OGREDataSet, enabling read-in of multiple datasets at once. External datasets show a wide range of file formats, structures, format, and naming conventions and are therefore not immediately ready for an overlap analysis. OGRE offers a growing number of built-in annotations for promoters, genes, CpG islands, SNPs, and TFBS. This is achieved by parsing functions that scan those datasets for duplicates, chromosome naming conventions, genome build and version differences. In addition, we provide instructions on how to process datasets from different origins. As illustrated in Fig. 1, the user is able to add and modify datasets within the Dataset module at any point. Integrated convenience functions allow resizing of input elements, making it possible to focus on specific regulatory regions like promoters or other up/downstream areas. For instance, dataset coordinates can be modified relative to the start/end positions, taking the DNA strand information into account (e.g. (−) 1200 bp from TSS). Next, overlap calculation is started by the Processing module, which operates on any supplied OGREDataSet and can be adjusted for multiple parameters like the minimum overlap required for two regions, type of overlap (i.e. full or partial), and strand-specific overlaps. The resulting hits, a pair of overlap** regions, is then further annotated by extracting genomic coordinates for each involved region pair, and used to generate tables containing comprehensive information underlying each overlap. In detail, the table contains genomic coordinates for both region pairs and for the overlap** region itself, length of overlap, and reports the overlapped nucleotide fraction with respect to the original input region. Some regions exhibit low overlap numbers whereby others, for example in promoter-TFBS or intergenic regions-SNP associations, typically show multiple overlaps. OGRE offers routines for extracting all elements overlap** a single region and thus identifies regions with many or few overlaps. Some genomic elements cluster around regulatory regions such as TFBSs upstream of genes. We therefore expect distinct coverage profiles, caused by an overlap enrichment at certain areas. To measure this, we divide all regions of a dataset of interest into 100 equally sized bins. In a next step we sum up all elements of a second dataset that fall into each of the bins. For a genes-TFBS dataset, this means every gene body is split into 100 bins, whereby the first bins start with the gene transcription start site and the last bins end with the gene transcription termination site. A matrix stores this information for all first dataset’s regions and a vector is defined containing the accumulated overlap coverage along the bins. A summary table displays informative statistics such as minimum, lower quantile, mean, median, upper quantile, and maximum number of overlaps per region and per dataset. The last module, Visualization, illustrates the summary table’s information as bar plots and generates histograms to display overlap distributions by grou** the number of overlaps into predefined bins. Chromosome, strand, start, and end coordinates of all datasets are then used to generate tracks for a local genomic visualization representing a user-defined genome window. Optionally, multiple layers of datasets, that were not directly part of the overlap calculation, can be displayed alongside the initially selected datasets. Appearance like colors, shapes, and labeling types can be adjusted and taken into account by the user. As an alternative exploration method, we implemented an interface to display overlap** regions on public genome browsers.
OGRE workflow. OGRE’s architecture is divided into three modules: Datasets (red), Processing (blue), and Visualization (green) Database access is interconnected with key processes, data generation, results generation, and visualization. Decision junctions (rhombus shaped) display the user’s options to influence number and type of datasets, dataset manipulation and visualization parameters
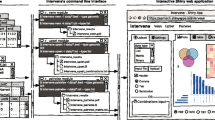
On the technical side, OGRE was programmed in R 4.1.0 [10] using the RStudio integrated development environment [11] and visualization is done with Shiny [12]. OGRE’s structure is displayed in Fig. 2, where input, processing, and output are interconnected with annotations from public databases. Most functionalities were implemented with the R base code and the use of additional packages, namely GenomicRanges [13] to calculate overlap between input regions and public annotations, DataTable [5: Table S2, Additional file 6: Table S3). In addition, OGRE reports the number of query regions, calculates the total number of annotation types found among query regions, regions with at least one regulatory element, and the average number of regulatory elements per query. Results are internally stored as data tables, which can be exported (Fig. 3B, Additional file 5: Table S2, Additional file 6: Table S3) and are in turn the input for visualization with ggplot2 and Gviz (Fig. 3A, C). Shiny is used to set up the convenient user interface SHREC. In more detail, we visualize input and output data with the ggplot2 [17] R package to create basic bar plots with information on the number of submitted queries/genes. Furthermore, the total and average number of subjects/regulatory elements found for every input is computed. OGRE makes extensive use of the DT [Full size image
Overlap** protein-coding genes in the human genome
Overlap** genes are defined as two or more genes sharing the same location by partially or entirely overlap** with each other. They exist mostly in compact genomes like those of virus and bacteria, however they are also found in the human genome. Their close genomic proximity results in sharing the same chromatin domains or compartments, which in turn leads to parallel regulation and transcription [26]. In a recent study Chen et al. [27] analyzed 19,200 well-annotated protein-coding genes and determined that 4951 (26%) of those overlapped with adjacent genes, with the biggest cluster containing 22 overlap** genes. In an effort to match the original analysis parameters, we used Ensembl’s GRCh38.p12 gene annotation release from April 2018 and filtered the dataset for protein-coding genes with description only. After running OGRE with this similar dataset of 19,308 protein coding genes, we report a total of 5407 (28%) genes overlap** with at least one other gene. These are 456 genes, 2% more than those identified by the authors. Both partial and complete overlaps were considered as hits, reported independently from DNA strand notation (i.e. forward and reverse), and were displayed using OGRE’s local visualization feature (Fig. 4C). On average, OGRE reported 0.3 overlaps per gene (min = 0, mean = 0.3, max = 22), with most overlaps found within the protocadherin gamma family cluster. Gene–gene overlaps tend to occur more often around gene start (5′) and end (3′), whereby overlaps around the center of the gene are less frequent (Fig. 4B).
Comparison to other tools
An overlap analysis between the user-defined regions and selected genomic annotations should be user-friendly, comprehensive, fully automated, be able to process multiple regions at once, provide annotation and detection for common regulatory elements e.g. CGI, TFBS, and promoters, and have the options to visualize and export results. The research community already offers a range of different algorithms and tools to predict or annotate genomic regions. We selected available tools with comparable features to OGRE and listed their performance among the different requirement categories (Table 1). Most tools are specialized on analyzing regions for a certain type of annotation and do not offer support for additional annotations. For example, INSECT, CiiiDER, and ConTra v3 feature prediction of TFBSs from position frequency matrices (PFM), iProEP focusses on the prediction of promoters and GaussianCpG on CGI identification. While these tools try to annotate regions based on predictions, Goldmine, regioneR, annotatr, and OGRE make use of already published annotations. We have benchmarked these packages for their overlap performance using microbenchmark [28], resulting in comparable runtimes (Goldmine 0.046 s, regioneR 0.040 s, annotatr 0.049 s, and OGRE 0.047 s) using identical input datasets, when calculating gene–gene overlap (Additional file 4: Table S1, Additional file 2: Fig. S1 and Additional file 1). All four tools report a total overlap of n = 10,014 by processing a dataset with 20,314 genes. regioneR and annotatr focus on the statistical analysis of genomic regions and do not offer a graphical user interface and genomic overlap plotting. OGRE on the other hand, excels by providing built-in annotations, processing of multiple input regions, and visualization of overlap at a genomic level, accessible through a convenient user interface (Fig. 3A).
Software tools with similar features were compared to OGRE on their capability to manage multiple input regions, built-in annotations, and visualize overlaps.