Abstract
Background
Rice black-streaked dwarf virus (RBSDV), a member of the genus Fijivirus within the family Reoviridae, causes severe damage to cereal crops in South East Asia. The protein P7-2, encoded by the second open reading frame of segment S7, is conserved among most plant-infecting fijiviruses, but its function is still obscure.
Results
In this study, P7-2 was used as bait in two-hybrid screens of a cDNA library expressing Zea mays proteins. It was found that there is a strong interaction between P7-2 and Z. mays SKP1 (SKP1Maize), a core subunit of the multicomponent SCF (SKP1/Cullin1/F-box/Rbx1) E3 ubiquitin ligase. The interaction was then confirmed in leaf epidermal cells of Nicotiana benthamiana by bimolecular fluorescence complementation assay. Further investigations indicated that P7-2 also interacts with SKP1 proteins from other plants, including Arabidopsis thaliana, N. benthamiana,Oryza sativa and Saccharum sinense. The C-terminal fragment of SKP1Maize (residues 97–176) and the middle fragment of P7-2 (residues 79–214) are necessary to sustain the interaction, while the C-terminal putative α-helix domain spanning residues 214–295 of P7-2 greatly facilitates the interaction. Agrobacterium-mediated transient suppression assay showed that P7-2 has no obvious activity to suppress local RNA silencing.
Conclusions
Taken together, our results indicated that RBSDV P7-2 can interact with SKP1 proteins from different plants. This is the first report linking a Fijivirus protein to a component of the ubiquitin proteasome system. P7-2 might be a potential F-box protein encoded by RBSDV and involved in the plant-virus interaction through ubiquitination pathway.
Similar content being viewed by others
Background
Rice black-streaked dwarf virus (RBSDV) is an insect- and plant-infecting agent that belongs to the genus Fijivirus in the family Reoviridae. The virus is transmitted propagatively via the small brown planthopper Laodelphax striatellus and causes rice black-streaked dwarf and maize rough dwarf diseases in southeast Asian countries [1–4]. Infected plants show typical stunting, darkening of leaves and waxy white tumors or black-streaked galls along the veins on the underside of leaf blades, sheaths and columns [1, 5].
The mature RBSDV virion is an icosahedral, double-layered particle with a diameter of 75–80 nm and consists of 10 genomic double-stranded RNA (dsRNA) segments (S1-S10) [6, 7]. Most genomic segments contain one open reading frame (ORF), except S5, S7 and S9, which are bicistronic. Six structural proteins (P1, P2, P3, P4, P8 and P10) and six nonstructural proteins (P5, P6, P7-1, P7-2, P9-1 and P9-2) are encoded by RBSDV genome. P1, P2, P3 and P4 are assigned to be the putative RNA-dependent RNA polymerase, a core protein, a putative cap** enzyme and the outer-shell B-spike protein of the virion, respectively, based on their deduced amino acid sequences and molecular masses [6–8]. P8 is the minor core capsid protein and possesses potential active transcriptional repression activity [9]. P10 is the main component of the outer shell of the viral particle and can self-interact to form trimers in solution [5, 10]. Both S7 and S9 have two non-overlap** ORFs and encode two nonstructural proteins. P7-1 is involved in the formation of tubular structures, and the secretory pathway and actomyosin motility system in plant hosts are required for its plasmodesmatal localization [5, Full size image
To determine the region responsible for P7-2-SKP1Maize interaction, four truncation derivatives were constructed, expressing AD-SKP11-67, AD-SKP11-101, AD-SKP11-128, and AD-SKP197-176, based on the domains of SKP1Maize. Their abilities to interact with P7-2 were investigated via YTH assay. Schematic representation of the different SKP1 truncation derivatives is shown in Figure 4B.
The YTH analysis indicated that AD-SKP11-67, AD-SKP11-101 and AD-SKP11-128 completely lost the capabilities to interact with P7-2, and no growth was observed, as well as the negative control. AD-SKP197-176, in which only 79 amino acids at the C-terminus were left, sustained the interaction with some decreased binding capability, compared to the whole SKP1 (Figure 4C). The results suggested that the C-terminal Skp1 domain is necessary to sustain the P7-2-SKP1 interaction, while N-terminal Skp1_POZ domain is helpful to stabilize the interaction in some degree. Our results are consistent with those of the crystal structure of the human F-box protein Skp2 bound to SKP1 [25]. The analysis of the crystal structure indicated that the last two helices of the BTB/POZ fold and the two helices of the C-terminal extension of Skp1 form the Skp2-binding site, corresponding to the SKP1Maize fragment spanning residues 111 to 176 exactly.
Both the middle and C-terminal region of P7-2 are greatly involved in P7-2-SKP1Maize interaction
The strong interaction of P7-2-SKP1Maize suggested that P7-2 might contain an F-box motif, a nonconserved N-terminally located ≈ 50 aa domain, through which FBPs binds SKP1. Thus, a protein secondary structure prediction of P7-2 was carried out, by the Jpred 3 and SCRATCH prediction server [26, 27], as well as its counterparts in other fijiviruses. The prediction showed that there are no specific secondary structures at the N-termini of these proteins, while the region at the C-termini, spanning residues 283–307 as for P7-2, is predicted with high probability to be in an α-helical conformation. A collection of P7-2 truncation derivatives that express BD-P7-225-309, BD-P7-244-309, BD-P7-279-309, BD-P7-21-295, BD-P7-21-287, BD-P7-21-214 and BD-P7-279-214 were constructed sequentially, and their capabilities to bind to SKP1Maize were tested. Schematic representation of P7-2 and other mutant derivatives is shown in Figure 5.
Map** of the crucial region of P7- 2 involved in the interaction with SKP1Maize. (A) Schematic representation of P7-2 and its truncations in the study. The full-length P7-2 (spanning residues 1 to 309) and its truncations are indicated by blue bars while the deleted regions by dashed lines. The predicted α-helix and the short sequence (LPFAELL) similar to F-box consensus sequence (LPxxI/L) were also indicated. The numbers denote the amino acid positions of the proteins. The ability of different P7-2 truncations to interact with SKP1Maize is indicated on the right (+, positive; -, negative). (B) The interaction between different P7-2 truncations and SKP1Maize in YTH assays. Yeast colonies expressing AD-SKP1Maize with BD-P7-225-309, BD-P7-244-309, BD-P7-279-309, and P7-2-LP(79–80)AA were able to grow well on the selective medium and turned blue. BD-P7-21-295 and BD-P7-21-214 showed decreased capability to interact with SKP1Maize. No growth was observed in the yeast expressing AD-SKP1Maize with BD-P7-21-287 and BD-P7-279-214.
The YTH analysis indicated that P7-225-309, P7-244-309 and P7-279-309 are able to bind SKP1Maize strongly, and the colonies expressing these combinations showed no obvious growth inhibition, compared to the intact P7-2. However, it is not the case when the truncations happened at the C-terminus. As is shown in Figure 5, P7-21-295, lost 14 residues at C-terminus, has a slightly decreased activity to interact with SKP1. When 21 residues were deleted, P7-21-287 showed a complete inability to attach SKP1. But when the deletion increased to 95 residues, P7-21-214, which lost the whole α-helix comprised in C-terminus, recovered the interaction with a more weakened capability (Figure 5B).
A short sequence LPFAELL (residues 79–85 in P7-2) was found to show some similarity to the F-box consensus sequence (LPxxI/L), in which LP are the most highly conserved residues [28, 29]. To investigate the effect of L79P80, a point mutant P7-2-LP(79–80)AA substituting two alanines for L79P80 was constructed. And P7-2-LP(79–80)AA was able to interact with SKP1Maize strongly (Figure 5B).
The results suggested that the middle fragment of P7-2, spanning residues from 79–214 is crucial to sustain the P7-2-SKP1Maize interaction. The putative α-helix region at the C-terminus, spanning residues 283–307, plays an important role to facilitate the interaction, and partial deletion of α-helix intends to induce some conformational changes so as to interrupt the association. L79P80 was not crucial during the interaction.
Agrobacterium-mediated transient suppression assay indicated P7-2 has no obvious activity to suppress local RNA silencing
P7-2-SKP1 interaction suggested that P7-2 might be involved in the UP pathway. Up to now, many viruses have evolved different strategies to hijack the ubiquitination/proteolysis machinery to enhance their replication or counteract the host defense, by targeting cellular factors for degradation [30, 31]. Considering the assumption that P7-2 might participate in virus multiplication and pathogenicity, its activity in suppressing post-transcriptional gene silencing (PTGS) was examined through Agrobacterium-mediated transient suppression assay.
As described previously [32], an Agrobacterium strain harboring a GFP expressing plasmid, pGDSmGFP, was mixed with another strain carrying the test plasmid, pGDS7-2His, and the mixture was co-infiltrated into leaves of N. benthamiana. The latter plasmid was able to express a His-tagged P7-2 protein. A construct expressing p19, the viral suppressor of RNA silencing (VSR) from Tomato bushy stunt virus (TBSV) was used as the positive control, while the empty vector as the negative control. The accumulation of GFP and P7-2 was detected by GFP antiserum and the specific anti-His-tag monoclonal antibodies, respectively.
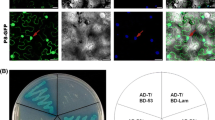
GFP fluorescence intensity in leaf regions co-infiltrated with pGDSmGFP and pGDS7-2His peaked at 3 days post-infitration (dpi), and declined rapidly over the next 2 days, as well as the negative combination of pGDSmGFP and pGD (Figure 6A). In contrast, when pGDSmGFP was co-infiltrated with pGDp19, the infiltrated patches displayed a bright green fluorescence even at 10 dpi. Western blot analysis confirmed the visual observation, for that the level of GFP protein accumulated were parallel to the intensity of GFP fluorescence (Figure 6B).
PTGS suppression of GFP by RBSDV P7- 2 in the N . benthamiana transient co- expression system. (A) Agro-infiltration of N. benthamiana with pGDSmGFP and pGD (left), pGDS7-2His (middle), and pGDp19 (right). Photographs were taken under long-wavelength UV light at 5 dpi. (B) Western blot analysis using GFP polyclonal antiserum. The intensity of the GFP signals was consistent with that of GFP fluorescence. Total proteins stained with Coomassie blue were used as the loading control.
The expression level of P7-2 accumulated to a higher level when the protein was co-expressed with p19, while no accumulation was detected when p19 was absent. The result suggested that P7-2 is able to be expressed in the leaves of N. benthamiana, but the protein exhibits no activity to suppress local gene silencing and is degraded rapidly without the existence of p19. To confirm our assumption, bacteria harboring pGDS7-2His was co-infiltrated with those containing pGDp19 and pGDSmGFP. GFP fluorescence and protein accumulation were compared with the control treatment co-infiltrated with the combination of pGD19 and pGDSmGFP. As we expected, P7-2 was expressed and detected in the infiltrated patch, and the fluorescence intensity and GFP accumulation were equivalent to the combination control. Taken together, P7-2 has no obvious local VSR activity.