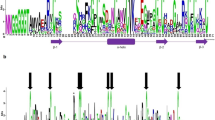
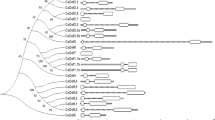
Abstract
Antimicrobial peptides (AMPs) are major components of innate immunity in plants and animals. AMP genes have significant intra- and interspecific polymorphism, the role of which is poorly understood. Previously, by high-throughput transcriptome sequencing of wheat plants, we identified defensin genes up-regulated upon infection with the pathogenic fungus Fusarium oxysporum and/or treatment with resistance inducers. In the present work, a bioinformatic search in NCBI databases for peptide homologs of these defensins was carried out using the sequences of their γ-cores, the sites of the molecules responsible for antimicrobial activity. DEFL1-16 homologs were identified in 95 species of angiosperms belonging to 48 families and 30 orders of monocotyledonous and dicotyledonous plants. The ubiquitous distribution of this defensin in angiosperms suggests its involvement not only in defense but also in other processes in flowering plants. Homologs of other defensins induced by infection were found only in plants of the Poaceae family, which suggests the existence of a Poaceae-specific defense mechanism associated with the expression of these defensins. Among the γ-core variants of wild plant defensins identified in the study, the peptides with better antimicrobial activity compared to wheat may be present, which are of considerable interest for the development of new antibiotics for medicine and agriculture.





Similar content being viewed by others
REFERENCES
Zasloff, M., Antimicrobial peptides of multicellular organisms, Nature, 2002, no. 415, pp. 389—395. https://doi.org/10.1038/415389a
Tam, J.P., Wang, S., Wong, K.H., and Tan, W.L., Antimicrobial peptides from plants, Pharmaceuticals, 2015, vol. 8, no. 4, pp. 711—757. https://doi.org/10.3390/ph8040711
Li, J., Hu, S., Jian, W., et al., Plant antimicrobial peptides: structures, functions, and applications, Bot. Stud., 2021, vol. 62, no. 1. https://doi.org/10.1186/s40529-021-00312-x
Lima, A.M., Azevedo, M.I.G., Sousa, L.M., et al., Plant antimicrobial peptides: an overview about classification, toxicity and clinical applications, Int. J. Biol. Macromol., 2022, vol. 214, pp. 10—21. https://doi.org/10.1016/j.ijbiomac.2022.06.043
Zou, F., Tan, C., Shinali, T.S., et al., Plant antimicrobial peptides: a comprehensive review of their classification, production, mode of action, functions, applications, and challenges, Food Funct., 2023, vol. 14, no. 12, pp. 5492—5515. https://doi.org/10.1039/d3fo01119d
Lazzaro, B.P., Zasloff, M., and Rolff, J., Antimicrobial peptides: application informed by evolution, Science, 2020, vol. 368, no. 6490. https://doi.org/10.1126/science.aau5480
Zhu, Y., Hao, W., Wang, X., et al., Antimicrobial peptides, conventional antibiotics, and their synergistic utility for the treatment of drug-resistant infections, Med. Res. Rev., 2022, vol. 42, no. 4, pp. 1377—1422. https://doi.org/10.1002/med.21879
Yount, N.Y. and Yeaman, M.R., Multidimensional signatures in antimicrobial peptides, Proc. Natl. Acad. Sci. U.S.A., 2004, vol. 101, no. 19, pp. 7363—7368. https://doi.org/10.1073/pnas.0401567101
Odintsova, T.I., Slezina, M.P., Istomina, E.A., et al., Defensin-like peptides in wheat analyzed by whole-transcriptome sequencing: a focus on structural diversity and role in induced resistance, Peer J., 2019, vol. 7. https://doi.org/10.7717/peerj.6125
Slezina, M.P., Istomina, E.A., Kulakovskaya, E.V., et al., The γ-core motif peptides of AMPs from grasses display inhibitory activity against human and plant pathogens, Int. J. Mol. Sci., 2022, vol. 23, no. 15. https://doi.org/10.3390/ijms23158383
https://blast.ncbi.nlm.nih.gov/Blast.cgi.
Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S., Improved prediction of signal peptides: SignalP 3.0, J. Mol. Biol., 2004, vol. 340, no. 4, pp. 783—795. https://doi.org/10.1016/j.jmb.2004.05.028
Kumar, S., Stecher, G., and Tamura, K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, no. 7, pp. 1870—1874. https://doi.org/10.1093/molbev/msw054
The Angiosperm Phylogeny Group, Chase, M.W., Christenhusz, M.J.M., et al., An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV, Bot. J. Linn. Soc., 2016, vol. 181, no. 1, pp. 1—20. https://doi.org/10.1111/boj.12385
Vekhov, V.N., Zostera morskaya Belogo morya (Common Eelgrass of the White Sea), Moscow: Mosk. Gos. Univ., 1992.
Zhizn’ rastenii (Plant Life), vol. 5, part 2: Tsvetkovye rasteniya (Flowering Plants), Takhtadzhyan, A.L., Ed., Moscow: Prosveshchenie, 1981.
Popov, A.P., Lekarstvennye rasteniya v narodnoi meditsine (Medicinal Plants in Alternative Medicine), Kiev: Zdorov’ya, 1967.
Usenko, N.V., Derev’ya, kustarniki i liany Dal’nego Vostoka (Trees, Shrubs, and Woody Vines of the Far East), Khabarovsk: Khabarovsk. Kn. Izd., 1984, pp. 110—111.
Li, P.-H., Shih, Y.-J., Lu, W.-C., et al., Antioxidant, antibacterial, anti-inflammatory, and anticancer properties of Cinnamomum kanehirae Hayata leaves extracts, Arab. J. Chem., 2023, vol. 16, no. 7, p. 104873. https://doi.org/10.1016/j.arabjc.2023.104873
Endress, P.K., Trochodendraceae, in Flowering Plants Dicotyledons: The Families and Genera of Vascular Plants, Berlin: Springer-Verlag, 1993, vol. 2, pp. 599—602. https://doi.org/10.1007/978-3-662-02899-5_74
Sun, Y., Deng, T., Zhang, A., et al., Genome sequencing of the endangered Kingdonia uniflora (Circaeasteraceae, Ranunculales) reveals potential mechanisms of evolutionary specialization, iScience, 2020, vol. 23, no. 5. https://doi.org/10.1016/j.isci.2020.101124
Li, C., Duan, C., Zhang, H., et al., Adaptative mechanisms of halophytic Eutrema salsugineum encountering saline environment, Front. Plant Sci., 2022, vol. 13. https://doi.org/10.3389/fpls.2022.909527
Dupin, S.E., Geurts, R., and Kiers, E.T., The non-legume Parasponia andersonii mediates the fitness of nitrogen-fixing rhizobial symbionts under high nitrogen conditions, Front. Plant Sci., 2020, vol. 10. https://doi.org/10.3389/fpls.2019.01779
Clarke, C.R., Timko, M.P., Yoder, J.I., et al., Molecular dialog between parasitic plants and their hosts, Annu. Rev. Phytopathol., 2019, vol. 57, pp. 279—299. https://doi.org/10.1146/annurev-phyto-082718-100043
Conran, J.G., Cephalotaceae, in Flowering Plants Dicotyledons: The Families and Genera of Vascular Plants, Berlin: Springer-Verlag, 2004, vol. 6, pp. 65—68. https://doi.org/10.1007/978-3-662-07257-8_7
Zhizn’ rastenii (Plant Life), vol. 6: Tsvetkovye rasteniya (Flowering Plants), Takhtadzhyan, A.L., Ed., Moscow: Prosveshchenie, 1982.
Li, C.J., Tsang, S.F., Tsai, C.H., et al., Momordica charantia extract induces apoptosis in human cancer cells through caspase- and mitochondria-dependent pathways, Evid. Based Complement. Alternat. Med., 2012, vol. 2012. https://doi.org/10.1155/2012/261971
Zhang, J., Hunto, S.T., Yang, Y., et al., Tabebuia impetiginosa: a comprehensive review on traditional uses, phytochemistry, and immunopharmacological properties, Molecules, 2020, vol. 25, no. 18. https://doi.org/10.3390/molecules25184294
Huang, W., Zhang, L., Columbus, J.T., et al., A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C4 photosynthesis, Mol. Plant, 2022, vol. 15, no. 4, pp. 755—777. https://doi.org/10.1016/j.molp.2022.01.015
Slezina, M.P., Istomina, E.A., Kulakovskaya, E.V., et al., Synthetic oligopeptides mimicking γ-core regions of cysteine-rich peptides of Solanum lycopersicum possess antimicrobial activity against human and plant pathogens, Curr. Issues Mol. Biol., 2021, vol. 43, no. 3, pp. 1226—1242. https://doi.org/10.3390/cimb43030087
Slezina, M.P., Istomina, E.A., Korostyleva, T.V., et al., Molecular insights into the role of cysteine-rich peptides in induced resistance to Fusarium oxysporum infection in tomato based on transcriptome profiling, Int. J. Mol. Sci., 2021, vol. 22, no. 11. https://doi.org/10.3390/ijms22115741
Stotz, H.U., Spence, B., and Wang, Y., A defensin from tomato with dual function in defense and development, Plant Mol. Biol., 2009, vol. 71, nos. 1—2, pp. 131—143. https://doi.org/10.1007/s11103-009-9512-z
Allen, A., Snyder, A.K., Preuss, M., et al., Plant defensins and virally encoded fungal toxin KP4 inhibit plant root growth, Planta, 2008, vol. 227, no. 2, pp. 331—339. https://doi.org/10.1007/s00425-007-0620-1
Mith, O., Benhamdi, A., Castillo, T., et al., The antifungal plant defensin AhPDF1.1b is a beneficial factor involved in adaptive response to zinc overload when it is expressed in yeast cells, Microbiol. Open, 2015, vol. 4, no. 3, pp. 409—422. https://doi.org/10.1002/mbo3.248
Funding
This work was supported by the Russian Science Foundation (grant no. 22-16-00010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Slezina, M.P., Istomina, E.A. & Odintsova, T.I. Biological Diversity of Genes Encoding Wheat Defensin Homologs. Russ J Genet 59, 1310–1319 (2023). https://doi.org/10.1134/S1022795423120116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795423120116