Abstract
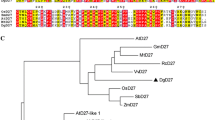
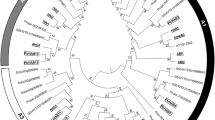
Strigolactones are a new class of plant hormones regulating shoot branching and symbiotic interactions with arbuscular mycorrhizal fungi. Studies of branching mutants in herbaceous plants have identified several key genes involved in strigolactone biosynthesis or signaling. The strigolactone signal is perceived by a member of the α/β-fold hydrolase superfamily, known as DWARF14 (D14). However, little is known about D14 genes in the woody perennial plants. Here we report the identification of D14 homologs in the model woody plant Populus trichocarpa. We showed that there are two D14 homologs in P. trichocarpa, designated as PtD14a and PtD14b that are over 95% similar at the amino acid level. Expression analysis indicated that the transcript level of PtD14a is generally more abundant than that of PtD14b. However, only PtD14a was able to complement Arabidopsis d14 mutants, suggesting that PtD14a is the functional D14 ortholog. Amino acid alignment and structural modeling revealed substitutions of several highly conserved amino acids in the PtD14b protein including a phenylalanine near the catalytic triad of D14 proteins. This study lays a foundation for further characterization of strigolactone pathway and its functions in the woody perennial plants.
Similar content being viewed by others
Introduction
Strigolactones (SLs) are a new class of plant hormones regulating shoot branching1,2 and symbiotic interactions with arbuscular mycorrhizal fungi3,4. In addition, SLs regulate many other processes in plant growth and development including primary root growth, lateral root formation, adventitious root formation, root hair development, seed germination, photomorphogenesis and nodulation (reviewed in references5,6,7,8,9,10,11), protonema branching in moss12, as well as responses to stresses13 and nutrient deficiency (reviewed in reference8). In the last decade, great progresses have been made to identify genes regulating the biosynthesis and signaling of SLs, in particular, by analyzing branching mutants in Arabidopsis, pea, rice and petunia (reviewed in references14,58. T1 transformants were selected using 20 μg/L hygromycin B. A minimum of 20 independent transgenic lines were selected for each transformation. Six independent transgenic lines were used for further studies. When T1 plants reached maturity, the number of primary rosette-leaf branches was counted.
RT-PCR analysis
To examine the expression of PtD14 genes in the Arabidopsis transgenic lines, total RNA was extracted from the rosette leaves of four-week-old plants using the Invisorb Spin Plant Mini Kit (Stratec Molecular). Two μg of total RNA were reversely transcribed to cDNA using Fermentas RevertAid Reverse Transcriptase (Thermo Scientific). PtD14-specific primers were used in PCR reactions. PCR amplification of Arabidopsis ACTIN2 served as a control in the analysis of Arabidopsis transgenic lines.
Quantitative RT-PCR
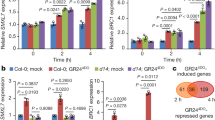
To examine the expression patterns of PtD14 genes, total RNA was extracted from various tissues and organs of Populus plants using the Spectrum™ Plant Total RNA isolation kit (Sigma). Two μg of total RNA were reversely transcribed to cDNA using Fermentas RevertAid Reverse Transcriptase. Quantitative RT-PCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific). Thermal cycling consisted of 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 15 s at 95 °C and 60 s at 60 °C. PtD14-specific primers (PtD14a: TTAGCCGAACGCTTTTCAACA and TTCCACAGTAGCTTTGCCACC; PtD14b: CTAAGAGGGATACTGGGCCT and TTCCACGGTATTTTCGCCAC) were used in the quantitative RT-PCR reactions. PCR amplification of Populus UBIQUITIN C served as a control for normalizing the relative transcript level. All PCR reactions were done with three technical replicates.
Yeast two-hybrid assay
The full-length open-reading frame of PtD14a, PtD14b and PtMax2a45 was each cloned into pENTR vector (Life technologies, CA). For the bait construct, the pENTR vector containing PtD14a or PtD14b was transferred into the pDEST32 destination vector by LR clonase-mediated reactions (Life technologies). For the prey construct, the pENTR vector containing PtMAX2a was transferred into the pDEST22 destination vector. One hundred ng of each plasmid of bait and prey construct was added into 100 μl of MaV203 competent yeast cells (Life technologies). For negative control, 100 ng of pDEST22 and pDEST32 empty vector was co-transformed with each other or with the counterpart of pDEST plasmid DNA. Co-transformation was performed by adding 600 μl of 40% PEG/1× LiAc to yeast cell and plasmid mixture followed by incubation at 30 °C for 30 min. After incubation, 35.5 μl of DMSO was added into the cell mixture to improve transformation efficiency. Then, the yeast cells were incubated for 20 min in a 42 °C water bath. Co-transformed yeast cell was centrifuged and the pellet was diluted in 1 ml of 0.9% NaCl. A total of 100 μl of diluted yeast cells was spread on SD plate deficient of Trytophane and Leucine (SD/-Trp/-Leu). Correctly co-transformed yeast cells were cultured in 2 ml of SD/-Trp/-Leu solution overnight at 28 °C. Cultured yeast cells were diluted up to 100 times with 0.9% NaCl. Fifteen μl of diluted yeast cells were dropped on SD plate deficient of Tryptophan, Leucine and Histidine (SD/-Trp/-Leu/-His) supplemented with 5 mM 3-Amino-1,2,4-triazole (3AT; Sigma-Aldrich, MO) or 5 mM of 3AT plus 5 μM GR24. The plates were incubated for 3 days at 28 °C. Yeast cells grown on SD plate were imaged with Canon power shot SX210 IS digital camera (Canon USA Inc., NY).
Protein degradation assay
Populus mesophyll protoplasts were isolated from P. tremula × alba clone 717-1B4 leaves as described previously46. A total of 30 μg of plasmid expressing 10× Myc-tagged PtD14 proteins was purified with Qiagen Plasmid Midi Kit and transfected into 200 μl of protoplasts (~2 × 105) using PEG-calcium mediated transfection method59. After 12 hr incubation at room temperature, GR24 was added into the transfected protoplast suspension to a final concentration of 5 μM and incubated for another 5 hr. Protoplasts were collected by centrifuging. Total protein was then extracted from protoplasts using 50 mM Tris-HCl (pH8.0), 100 mM NaCl, 10 mM EDTA (pH 8.0), 1% SDS, 1 mM PMSF and protease inhibitor (Sigma). After centrifuging, the supernatant was collected and protein concentration was determined by Bradford method. Protein extracts were mixed with SDS loading buffer (60 mM Tris-HCl pH 8.0, 1% SDS, 10% glycerol, 20 mM DTT) and denatured by boiling for 8 min. To detect 10× Myc-tagged PtD14 proteins with western blotting, 1 μg of total protein was separated in 10% SDS-PAGE gel and transferred to PVDF membrane. Membranes were then probed with anti-Myc antibody (1:4000; Abgent), detected with ECL reagent (Thermo) and imaged with CCD imager (Bio-rad). In parallel, same amount of protein extracts were separated in 10% SDS-PAGE gel and stained with ProteoSilver Silver Stain Kit (Sigma). One protein band existing in all samples (~80 kDa) was selected to demonstrate equal protein loading.
Additional Information
How to cite this article: Zheng, K. et al. Characterization of DWARF14 Genes in Populus. Sci. Rep. 6, 21593; doi: 10.1038/srep21593 (2016).
References
Gomez-Roldan, V. et al. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008).
Umehara, M. et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008).
Akiyama, K., Matsuzaki, K. & Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005).
Besserer, A. et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4, 1239–1247 (2006).
Koltai, H. Strigolactones are regulators of root development. New Phytol. 190, 545–549 (2011).
Seto, Y., Kameoka, H., Yamaguchi, S. & Kyozuka, J. Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol. 53, 1843–1853 (2012).
Brewer, P. B., Koltai, H. & Beveridge, C. A. Diverse roles of strigolactones in plant development. Mol Plant 6, 18–28 (2013).
Czarnecki, O., Yang, J., Weston, D. J., Tuskan, G. A. & Chen, J. G. A dual role of strigolactones in phosphate acquisition and utilization in plants. Int. J. Mol. Sci. 14, 7681–7701 (2013).
Mason, M. G. Emerging trends in strigolactone research. New Phytol. 198, 975–977 (2013).
Rasmussen, A., Depuydt, S., Goormachtig, S. & Geelen, D. Strigolactones fine-tune the root system. Planta 238, 615–626 (2013).
Ruyter-Spira, C., Al-Babili, S., van der Krol, S. & Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 18, 72–83 (2013).
Proust, H. et al. (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539 (2011).
Ha, C. V. et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 111, 851–856 (2014).
Beveridge, C. A. & Kyozuka, J. New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13, 34–39 (2010).
**e, X. N., Yoneyama, K. & Yoneyama, K. The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117 (2010).
Domagalska, M. A. & Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221 (2011).
Wang, Y. H. & Li, J. Y. Branching in rice. Curr. Opin. Plant Biol. 14, 94–99 (2011).
Tsuchiya, Y. & McCourt, P. Strigolactones as small molecule communicators. Mol. Biosyst. 8, 464–469 (2012).
de Saint Germain, A., Bonhomme, S., Boyer, F. D. & Rameau, C. Novel insights into strigolactone distribution and signalling. Curr. Opin. Plant Biol. 16, 583–589 (2013).
Zheng, Z., Germain Ade, S. & Chory, J. Unfolding the mysteries of strigolactone signaling. Mol. Plant 7, 934–936 (2014).
Bennett, T. & Leyser, O. Strigolactone signalling: standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22, 7–13 (2014).
Seto, Y. & Yamaguchi, S. Strigolactone biosynthesis and perception. Curr. Opin. Plant Biol. 21, 1–6 (2014).
Waldie, T., McCulloch, H. & Leyser, O. Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79, 607–622 (2014).
Stirnberg, P., van de Sande, K. & Leyser, H. M. O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141 (2002).
Sorefan, K. et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17, 1469–1474 (2003).
Booker, J. et al. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14, 1232–1238 (2004).
Booker, J. et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8, 443–449 (2005).
Lin, H. et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525 (2009).
Waters, M. T., Brewer, P. B., Bussell, J. D., Smith, S. M. & Beveridge, C. A. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159, 1073–1085 (2012).
Arite, T. et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 (2009).
Waters, M. T. et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 (2012).
Hamiaux, C. et al. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012).
Nakamura, H. et al. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4, 2613 (2013).
Jiang, L. et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013).
Zhou, F. et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 (2013).
Zhao, J. et al. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 55, 1096–1109 (2014).
Chevalier, F. et al. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150 (2014).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–1186 (2012).
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12, 7–8 (2015).
Zhao, L. H. et al. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 (2013).
Kagiyama, M. et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 (2013).
Gaiji, N., Cardinale, F., Prandi, C., Bonfante, P. & Ranghino, G. The computational-based structure of Dwarf14 provides evidence for its role as potential strigolactone receptor in plants. BMC Res. Notes 5, 307 (2012).
Czarnecki, O. et al. Characterization of MORE AXILLARY GROWTH genes in Populus. PLoS ONE 9, e102757 (2014).
Guo, J. et al. Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS ONE 7, e44908 (2012).
Schranz, M. E., Mohammadin, S. & Edger, P. P. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15, 147–153 (2012).
Campanella, J. J., Bitincka, L. & Smalley, J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4, 29 (2003).
Dereeper, A., Audic, S., Claverie, J. M. & Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8 (2010).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 (Web Server issue), W465–469 (2008).
van Dongen, S. Graph clustering via a discrete uncoupling process. Siam J. Matrix Anal. Appl. 30, 121–141 (2008).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Zhang, Y. Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins 82 (Suppl 2), 175–187 (2014).
Laskowski, R. A. PDBsum new things. Nucleic Acids Res. 37, D355–D359 (2009).
Roberts, E., Eargle, J., Wright, D. & Luthey-Schulten, Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7, 382 (2006).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996).
Nakagawa, T. et al. Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in Transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100 (2007).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protoc 2, 1565–1572 (2007).
Acknowledgements
This work was supported by the Plant-Microbe Interfaces Scientific Focus Area in the Genomic Science Program, the Office of Biological and Environmental Research in the U.S. Department of Energy Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the United States Department of Energy under contract DE-AC05-00OR22725. K.Z. and X.W. were partially supported by visiting scholarships from the China Scholarship Council. J.Y. was partially supported by a visiting scholarship from the Chinese Academy of Sciences (Grant Number: 201019).
Author information
Authors and Affiliations
Contributions
K.Z., X.W. and J.-G.C. conceived the experiment, K.Z., X.W., D.A.W., H.-B.G., M.X., Y.Y. and J.Y. conducted the experiments, K.Z., X.W., D.A.W., H.-B.G., M.X., Y.Y., J.Y., S.W., D.A.J., H.G., W.M., G.A.T. and J.-G.C. analyzed the results. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zheng, K., Wang, X., Weighill, D. et al. Characterization of DWARF14 Genes in Populus. Sci Rep 6, 21593 (2016). https://doi.org/10.1038/srep21593
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep21593
- Springer Nature Limited
This article is cited by
-
Genome-wide identification and analysis of the SUPPRESSOR of MAX2 1-LIKE gene family and its interaction with DWARF14 in poplar
BMC Plant Biology (2023)
-
Strigolactones: diversity, perception, and hydrolysis
Phytochemistry Reviews (2023)
-
Strigolactones in Plants: From Development to Abiotic Stress Management
Journal of Plant Growth Regulation (2023)
-
Molecular mechanism of lateral bud differentiation of Pinus massoniana based on high-throughput sequencing
Scientific Reports (2021)
-
Mechanistic Insights into Strigolactone Biosynthesis, Signaling, and Regulation During Plant Growth and Development
Journal of Plant Growth Regulation (2021)