Abstract
Self-supervised representation learning (SSL) on biomedical networks provides new opportunities for drug discovery; however, effectively combining multiple SSL models is still challenging and has been rarely explored. We therefore propose multitask joint strategies of SSL on biomedical networks for drug discovery, named MSSL2drug. We design six basic SSL tasks that are inspired by the knowledge of various modalities, inlcuding structures, semantics and attributes in heterogeneous biomedical networks. Importantly, fifteen combinations of multiple tasks are evaluated using a graph-attention-based multitask adversarial learning framework in two drug discovery scenarios. The results suggest two important findings: (1) combinations of multimodal tasks achieve better performance than other multitask joint models; (2) the local–global combination models yield higher performance than random two-task combinations when there are the same number of modalities. We thus conjecture that the multimodal and local–global combination strategies can be treated as the guideline of multitask SSL for drug discovery.
Similar content being viewed by others
Main
Drug discovery is an important task for improving the quality of human life; however, it is an expensive, time-consuming and complicated process that has a high chance of failure1,2. To improve the efficiency of drug discovery, a great number of researchers are devoted to develo** or leveraging deep learning to speed up its intermediate steps, such as molecular property predictions3,4, drug–target interaction (DTI) predictions5,6,7,8,9,10,11 and drug–drug interaction (DDI) predictions12,13. A key advantage to these methods is that deep learning algorithms can capture the complex nonlinear relationships between input and output data14.
Deep learning techniques have in the past few years gradually emerged as a powerful paradigm for drug discovery. Most deep learning architectures such as convolutional15 and recurrent16 neural networks operate only on regular grid-like data (for example, 2D images and text sequences), and are not well suited for graph data (for example, DDI and DTI networks); however, in the real world, biomedical data are often formed as graphs or networks. In particular, biomedical heterogeneous networks (BioHNs) that integrate multiple types of data source are used extensively for life-science research. This is intuitive as BioHNs are well suited for modelling complex interactions in biological systems. For example, BioHNs incorporating DDIs, DTIs, protein–protein interactions (PPIs) and protein–disease associations can naturally simulate the 'multi-drug, multi-target, multi-disease’ biological processes within the human body17. In the context of biomedical networks applications, graph neural networks (GNNs)18,19,20—deep learning architectures specifically designed for graph structure data—are used to improve drug discovery. Past works21,22,23,24 have used GNNs to generate the representation of each node in BioHNs, and formulate drug discovery as the node- or edge-level prediction problems. Such graph neural network-based drug discovery approaches have exhibited high-precision predictions, but most existing methods heavily depend on the size of the training samples; that is, only large-scale training samples can help models achieve great performance. The performance drastically changes with the variation in the size of the training sample. Unfortunately, data labelling is expensive and time-consuming. These graph-based deep learning models that rely on large-scale labelled data may therefore not be satisfactory in real drug development scenarios.
Self-supervised representation learning (SSL) is a promising paradigm for solving the above issues. In SSL, deep learning models are trained via pretext tasks, in which supervision signals are automatically extracted from unlabelled data without the need for manual annotation. Self-supervised representation learning aims to guide models towards generating generalized representations to achieve better performance on various downstream tasks. Following the immense success of SSL on computer vision25,26 and natural language processing27,28, SSL models built upon BioHNs are attracting increasing attention and have been successfully applied to drug discovery29,30,31,32. Unfortunately, most existing methods often design a single SSL task to train GNNs for drug discovery, thus leading to a built-in bias towards the single task while ignoring the multiperspective characteristics of BioHNs. To cope with the potential bottleneck in single-task-driven SSL applications, there have been a few attempts at leveraging multiple SSL tasks for facilitating the performance of drug discovery33,34,35. These methods aim to integrate the advantages of various types of SSL tasks via multitask learning paradigms; however, most past approaches train GNNs according to a fixed joint strategy involving multiple tasks and do not focus on the differences between various multitask combinations. At the same time, the determination of which combination strategies can generate the most effective improvements has rarely been explored. It is therefore important to pay attention to the choice of multitask combination strategies in SSL approaches. Multitask SSL methods built on BioHNs for drug discovery are still in the initial stages, and more systematic studies are urgently needed.
We propose multitask joint strategies of SSL on biomedical networks for drug discovery (MSSL2drug) to address the aforementioned problems. Inspired by three modality features (structures, semantics and attributes in BioHNs), six self-supervised tasks are developed to explore the impact of various SSL models on drug discovery. Next, fifteen multitask joint strategies are evaluated via a graph-attention-based multitask adversarial learning model in two drug discovery scenarios. We find that combinations of multimodal tasks exhibit superior performance to other multitask strategies. Another interesting conclusion is that the local–global combination models tend to yield better results compared with random task combinations when there are the same number of modalities.
Result
Overview of MSSL2drug
We demonstrate the schematic workflow of MSSL2drug in Fig. 1. First we construct a BioHN that integrates 3,046 biomedical entities and 111,776 relationships. Second, we develop six self-supervised tasks based on structures, semantics and attributes in the BioHN (Fig. 1b). These self-supervised tasks guide graph attention networks (GATs) to generate representations from different views in the BioHN. More importantly, we develop fifteen kinds of multitask combinations and a graph-attention-based multitask adversarial learning framework (Fig. 2) to improve representation quality. Finally, the different single- and multitask SSL representations are fed into the multilayer perceptron (MLP) for predicting DDIs and DTIs. We can draw two important findings on the basis of the experiment results: (1) the combinations of multimodal SSL tasks achieve a state-of-the-art drug discovery performance; (2) the joint training of local and global SSL tasks is superior to the random combinations of two SSL tasks when there are the same number of modalities.
a, The BioHN is constructed. b,c, Six self-supervised tasks are developed (b), which guide GATs to generate representations from different views in the BioHN (c). e, Representation vectors are generated. d,f, Fifteen kinds of multitask combinations (d) and a graph-attention-based multitask adversarial learning framework (f) are developed. g, The different single- and multitask SSL representations are fed into the MLP. h, The two important findings from MSSL2drug results. All circles, quadrangles and pentagons denote the drugs, proteins and diseases in a BioHN, respectively. The solid lines are the relationships among the biomedical entities in a BioHN. The red nodes represent the randomly selected vertices or node pairs in each of self-supervised task. The red solid lines in the edge type masked prediction (EdgeMask) and bio-path classification (PathClass) modules represent the randomly selected edges or paths, respectively. The red dashed curves in the pairwise distance classification (PairDistance) module represent the measurements of the shortest paths between biomedical entities. The red solid curves in the node similarity regression (SimReg) and node similarity contrast (SimCon) modules represent the measurements of the similarities between biomedical entities. ClusterPre and PairDistance denotes clustering coefficient prediction and a pairwise distance classification, respectively.
For each epoch, we randomly select a SSL task, tn, from multitask combinations. The corresponding private and shared GAT models generate the task-specific (Rn) and common (Rs) representations, respectively; Rn and Rs are concatenated, and then fed into the MLP-based predictor of SSL task tn. The Rs values are fed into the MLP-based discriminator to predict which type of task the shared representation vectors come from. The parameters of current private and shared GAT models are updated by back-propagation based on the loss values from a SSL task predictor and discriminator, respectively. Finally, the parameters of the current shared model are assigned to all of the other shared models. We therefore attain n private GAT models and shared GAT models with same parameters after multitask SSL training. In other words, MSSL2drug generates the private representations by all private GATs and the shared representation by an arbitrary shared model.
Performance of a single-task-driven SSL
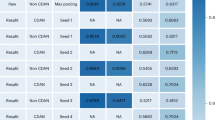
PairDistance and PathClass achieve relatively high results in a single-task-driven SSL for drug warm-start predictions (Fig. 3). Based on a Student’s t-test on the DTI and DDI results (Supplementary Section 1), we find that they considerably outperform ClusterPre and EdgeMask (P-value < 0.05). Another aspect to note is that SimCon peforms better than SimReg. These results suggest that the global-information-driven SSL approaches are superior to the local-information-based SSL; an earlier study36 also made a similar finding. In addition, we find that attribute-weak constraint-based SSL tasks outperform strong constraint-based models.
a–f, Results for DDI-AUROC (a), DDI-AUPR (b), DTI-AUROC (c), DTI-AUPR (d), DDI-AUROC and DDI-AUPR (e), and DTI-AUROC and DTI-AUPR (f). The area under precision recall (AUPR) and area under receiver operating characteristic (AUROC) curves are used for the evaluation metrics. The mean and s.d. values are calculated across ten results.
Local–global tasks achieve superior performance
In this experiment, first, eleven two-task combination models are divided into two categories: single- and double-modality combinations. It is noted that we design self-supervised tasks inspired by the knowledge of various modalites, including structures, semantics and attributes in BioHNs; therefore, there are up to three single-modality combination models (Fig. 4b). Second, we compare the performance of two-task models with the same number of modalities. The results in Fig. 4 suggest that joint training of local and global SSL tasks (that is, EdgeMask–PairDistance, ClusterPre–PathClass, ClusterPre–PairDistance and EdgeMask–PathClass) tends to lead to higher performance than random combinations of two SSL tasks when there are the same number of modalities (we further investigate the difference among various methods in Supplementary Section 2). We therefore conjecture that the local–global combination strategies can be regarded as an effective guideline for multitask SSL to drug discovery.
a,b, Heatmap of two-task combinations in the double- (a) and single-modality (b) combinations. The results are normalized to [0,1] along the x-axis by the min–max normalization technique. The redder (bluer) squares denote the greater (smaller) value. The shaded area denotes the combinations of global and local SSL tasks.
Multimodal tasks achieve best performance
The results in Fig. 5 show an interesting scenario: the growth of modalities leads to a substantial performance improvement (P-value < 0.05) for drug discovery (further results and Student’s t-test analyses can be found in Supplementary Section 3). These results suggest that combinations of multimodal tasks can achieve superior performance for drug discovery. We therefore conjecture that the multimodal combination strategy can be regarded as a potential guideline for multitask SSL for drug discovery.
a, Results for two-task combinations with different modalities. b,c, Results for multitask combinations from attributes (b) and structures (c) to multimodality. The total number of tasks and modalities in each multitask combination are denoted by T and M, respectively. The mean and s.d. values are calculated across ten results.
Performance of MSSL2drug on cold-start predictions
For the cold-start drug prediction scenarios, the results of DDI and DTI predictions are generated by six basic SSL tasks and fifteen kinds of multitask combinations. These results are straightforward and effective demonstrations that global-information- and attribute-weak constraint-based SSL models can achieve better performance than local information and attribute strong constraint-based SSL. More importantly, these results verify that multimodal and local–global combination strategies can achieve state-of-the-art prediction drug discovery performance (detailed analyses can be found in Supplementary Section 4).
Performance validation of MSSL2drug on external dataset
To demonstrate the robustness of MSSL2drug, it is used for Luo’s dataset6 and evaluated by warm- and cold-start predictions with different splitting ratios. The detailed setting and result analysis can be found in Supplementary Section 5. The results on warm-start predictions suggest that the multimodal and local–global combination strategies still conducive to improving the performance of drug discovery on Luo’s dataset. The performance of all SSL models on small training data and cold-start predictions is reduced, because the volume of training set is reduced. However, we find the same performance distribution, that is, the multimodal and local–global combination strategies tend to generate better prediction performance. The results on different splitting ratios further demonstrate that MSSL2drug has the high robustness and generalization.
Performance comparisons
To demonstrate superiority of MSSL2drug, PairDistance–EdgeMask–SimCon is compared with six state-of-the-art methods, including deepDTnet37, MoleculeNet38, KGE_NFM5, DTINet6, DDIMDL39 and DeepR2cov31. On the constructed biomedical network data shown in Table 1, we find that PairDistance–EdgeMask–SimCon is superior to the six baselines. PairDistance–EdgeMask–SimCon still outperforms other methods on Luo’s dataset for warm-start predictions, as shown in Supplementary Table 12. We also compare MSSL2drug with Laplacian Eigenmaps40, Graph Factorization41, DeepWalk42, MF2A43 and MIRACLE44. The results suggest that MSSL2drug can achieve higher performance on different datasets and scenarios. (see Supplementary Sections 6 and 16 for more details). We compare the run-time and parameter sizes in Supplementary Section 12.
The MSSL2drug and six baselines were also evaluated under different splitting ratios between the training and test sets (Supplementary Fig. 6). We observe that the performance of all methods are reduced when there are only few training samples. In particular, when the ratio of training:test sets is 5:95 or 10:90, all methods achieve poor results for DDI and DTI predictions. An interesting finding is that the performance of MSSL2drug is without much fluctuation, and superior to baselines for different volumes of training sets. These results suggest that most existing methods are prone to be influenced when applying to a small dataset, whereas MSSL2drug can partly overcome this limitation.
Application in drug repositioning for COVID-19
As coronavirus disease 2019 (COVID-19) has recently posed a global health threat, we apply MSSL2drug to drug repositioning for COVID-19, aiming to discover agents that inhibit IL-6 and therefore block the excessive inflammatory response in patients. Based on PubMed publications, clinical studies, molecular docking and molecular dynamics, we find that most of the predicted drugs may be able to inhibit the release of IL-6. More importantly, vandetanib (KD = 28.6 μM) and pazopanib (KD = 20.7 μM) can bind to IL-6 with high affinity as measured by a surface plasmon resonance assay45 (see Supplementary Section 7 for detailed descriptions); however, it is necessary to further validate via standard and systematic experiments whether there are indirect relationships or physical interactions between these drugs and IL-6. All of the predicted drugs must also be validated in preclinical experiments and randomized clinical trials before being administered to patients.
Impact of key components on performance
Key component analyses in SSL tasks
-
Selection of centrality measurements in ClusterPre: Compared with degree and eigenvector centrality46, the clustering coefficient-based SSL model47 achieves higher results (Supplementary Section 8.1). A possible explanation for this result is that the clustering coefficients are not only extract the distribution of neighbouring nodes, but also the triangle (loops of order 3) structures48 in networks.
-
Division of major class in PairDistance: The results suggest that dividing 4-hop and higher-hop node pairs into a major class achieves better performance compared with 3-hop and 5-hop (Supplementary Section 8.2). This phenomenon is consistent with the finds in S2GRL (ref. 49).
-
Length of meta path in PathClass: We observe that selecting meta paths with lengths 4 is contribute to the performance of PathClass when compared to other length paths. Previous studies have made a similar finding50,51. The details can be found in Supplementary Section 8.3.
-
Selection of similarity measurement in SimCon: We find that different similarity measurements bring the marginal improvements or reductions to SimCon (Supplementary Section 8.4). A possible explanation for this result is that SimCon only requires to distinguish the similarity distributions between node pairs, thus reducing the dependence on similarity measurements.
-
Ablation analyses of PairDistance–EdgeMask–SimCon: We further suggest that integrating multimodal and local–global task is beneficial to improve performance of drug discovery. In PairDistance–EdgeMask–SimCon, the contribution of SimCon is relatively lower than EdgeMask and PairDistance to some extent (Supplementary Section 8.5).
Component analyses in multitask learning framework
In this section we evaluate the respective contributions of the adversarial training (ADL) strategy and orthogonality constraint (ORC) mechanism to MSSL2drug (a detailed description and the results are provided in Supplementary Section 9). We find that MSS2drug achieves superior performance when compared with the ADL and ORC models. In other words, MSS2drug integrating ADL and ORC is beneficial to improve performance of drug discovery. We also find that the contribution of ORC is higher than ADL to some extent. Furthermore, each task is trained by turn in a stochastic manner. The random task orders tend to be more robustness and reliability than the fixed task orders; however, we conjecture that using the prior or domain knowledge to set a specific order may contribute to the improvement of multitask models.
High-quality representation analyses
In this experiment, the representations from MSSL2drug are fed into Random Forest52 and support vector machine53 for drug discovery predictions. We find that using support vector machine and Random Forest still achieves great performance for DDI and DTI predictions (Supplementary Section 10). These results suggest that MSSL2drug can generate the high-quality representations that can keep the inherent nature of BioHNs, thus improving the performance of drug discovery.
Dataset contamination analyses
We remove the DTI of test set from the SSL stage to understand how much influence the data contamination in SSL has on DTI predictions. The results suggest that the data contamination in SSL does not cause a substantial change in the performance of MSSL2drug. In other words, MSSL2drug is relatively insensitive to data contamination (Supplementary Section 11).
Discussion
Self-supervised representation learning on BioHNs has recently emerged as a promising paradigm for drug discovery. We therefore aim to explore a combination strategy of multitask self-supervised learning on BioHNs networks for drug discovery. Based on six self-supervised learning tasks, we find that global-knowledge-based SSL models outperform local-information-based SSL models for drug discovery. This is intuitive and understandable as global view-based SSL tasks can capture the complex structures and semantics that cannot be naturally learned by local SSL models. We also find that attribute-weak constraint-based SSL tasks are superior to strong constraint-based models. This may be attributed to the fact that the similarity scoring functions are handcrafted and unable to accurately reflect the similarities among nodes in the original feature space. Unfortunately, the node similarity regression tasks arbitrarily fit node similarity values of node pairs. By contrast, similarity contrast tasks reduce the dependence on the original feature similarity values.
More importantly, fifteen kinds of multiple task combinations are evaluated by a graph-attention-based multitask adversarial learning model for drug discovery. These results suggest that the joint training the global and local tasks can achieve the relatively high prediction performance when there are the same number of modalities. By contrast, combining the tasks with great performance does not necessarily lead to better performance than other multitask combinations for drug discovery. This is intuitive as there may be some conflicts and redundancies in the random combinations of SSL tasks; however, the combinations of global and local SSL models enable GNNs to leverage complementary information in BioHNs. To be specific, the local graph SSL models can capture the features within node itself or its first-order neighbours, but ignore the bird’s-eye view of the node position in BioHNs. Fortunately, global SSL models can learn the dependencies among long-range neighbourhoods, thus compensating the shortcomings of local SSL tasks. Simultaneously, an interesting finding is that combination models with multimodal tasks tend to generate best performance. This is because the combinations of multimodal tasks can capture multi-view information including structure, semantic and attribute features in BioHNs. The multimodal SSL models allow for knowledge transfer across multiple views and attain a deep understanding of natural phenomena in BioHNs. For a given SSL task, there are different levels of contributions in different multitask combinations. Generally, if a SSL task can bring new modality information to multitask models, it will generate the relatively greater contributions. Furthermore, if a local (global) information-driven SSL task is added to global (local) information-driven SSL tasks, it tends to bring a high performance improvement. The multimodal and local–global combination strategies may be prioritized when develo** multitask SSL for drug discovery. In other words, you can yourself design multitask SSL models according to the multimodal and local–global combination strategies when you want to use MSSL2drug for drug discovery. On the other hand, you can also directly use PairDistance–EdgeMask–SimCon for drug discovery, because it integrates the multimodal and local–global SSL tasks, and achieves best performance.
In the application of deep learning, when there is a relative scarcity of labelled data, it is easy to cause the overfitting problems, which exhibit a low testing performance even though its training performance is larger54. Fortunately, a great number of studies have suggested that multitask learning techniques can greatly reduce the risk of overfitting55,56,57,58,68 and BioGRID69. Each protein name is transferred into an Entrez ID (https://www.ncbi.nlm.nih.gov/gene) via the NCBI (https://www.ncbi.nlm.nih.gov/). Drug–disease associations are attained via the fusion of the drug indications in the repoDB70, DrugBank (v.4.3) and DrugCentral databases71. Disease–protein associations are collected from two databases, including the Online Mendelian Inheritance in Man database72 and the Comparative Toxicogenomics database73. The disease names are standardized according to Unified Medical Language System vocabularies74, and mapped to the MedGen ID (https://www.ncbi.nlm.nih.gov/medgen/) based NCBI database. BioHN in this work includes less information profiles than the dataset deepDTnet. Finally, the BioHN contains 3,046 nodes and 111,776 relationships (Supplementary Table 26). There are 1,894 proteins, 721 drugs, and 4,978 drug–protein interactions in the BioHN. The ratio of DTI labels is 0.003 ≈ 4,978/(721 × 1,894). Similarly, there are 66,384 DDIs in the BioHN; the ratio of DDI label is thus 0.256 ≈ 66,384/(721 × 720 × 0.5). In other words, there are sparse labels for DDI and DTI predictions. We therefore propose MSSL2drug, which explores multitask joint strategies of SSL on biomedical networks for drug discovery.
Basic self-supervised learning tasks
Multimodal information such as structures, semantics and attributes in BioHNs provides unprecedented opportunities for designing advanced self-supervised pretext tasks. Hence we develop six self-supervised tasks on the basis of the multimodal information contained in BioHNs for drug discovery.
Structure-based SSL tasks
The first direct choice for constructing SSL tasks is the inherent structure information contained in BioHNs. For a given node, self-supervision information is not only limited to itself or local neighbours, but also includes a bird’s-eye view of the node positions in a BioHN. We therefore design a clustering coefficient prediction (ClusterPre) task that captures local structures and a pairwise distance classification (PairDistance) task that reflects the global structure information in BioHNs.
ClusterPre. In this pretext task, we use GATs to predict the clustering coefficient47 of each node in the BioHNs. The ClusterPre SSL task aims to guide GATs to generate low-dimensional representations that preserve the local structure information in BioHNs. In ClusterPre, the loss function adopts the mean squared error (Supplementary Section 14.1).
PairDistance. We develop PairDistance, which is not limited to a node itself and its local neighbourhoods; it also takes global views of a BioHN. Similar to S2GRL (ref. 17a is equivalent to Supplementary Fig. 17b.
For a given node, different SSL tasks in different epochs guide the shared GAT to capture the features with itself task property. Self-supervised training in different epochs can thereofre be treated as the adversarial learning process, that is, each SSL task encourages shared GAT to generate task-specific representations. After sufficient training, the shared GATs reach a point, at which it integrates the property of different tasks; the shared feature space thus simply contains common information. By contrast, the private GAT model generates task-specific representations to make accurate SSL predictions.
Initialization features
In MSSL2drug, the initialization features of each node and adjacency matrixes of BioHNs are fed into GATs to perform training and test. Here we take an example to describe the process of feature initialization, as shown in Supplementary Fig. 18. There are three key steps to generate the initialization features. For each given node, its neighbours are divided into three categories (drugs, proteins and diseases).
Step 1: Counting the number of neighbours in each class, \(X = \left\{ {x_1,x_2, \cdots ,x_N} \right\}\), \(Y = \left\{ {y_1,y_2, \cdots ,y_N} \right\}\) and \(Z = \left\{ {z_1,z_2, \cdots ,z_N} \right\}\), where N is the total number of nodes. For instance, for given node 1, \(x_1 = {{{\mathrm{1}}}},y_1 = {{{\mathrm{2}}}},z_1 = {{{\mathrm{1}}}}\), the sum of \(x_1,y_1,z_1\) is its degree (that is, the number of its neighbours), as shown in the first row in Supplementary Fig. 18b;
Step 2: Converting X, Y and Z to matrixes \({{{\mathbf{X}}}} = \left\{ {\overrightarrow{u} _1,\overrightarrow{u} _2, \cdots ,\overrightarrow{u} _N} \right\}\), \({{{\mathbf{Y}}}} = \left\{ {\overrightarrow{v} _1,\overrightarrow{v} _2, \cdots ,\overrightarrow{v} _N} \right\}\) and \({{{\mathbf{Z}}}} = \left\{ {\overrightarrow{g} _1,\overrightarrow{g} _2, \cdots ,\overrightarrow{g} _N} \right\}\) by one-hot encoding technologies (https://www.educative.io/blog/one-hot-encoding);
Step 3: Generating initialization feature matrix \({{{\mathbf{F}}}} = \left\{ {\overrightarrow{u} _1||\overrightarrow{v} _1||\overrightarrow{g} _1,} \right.\) \(\left. {\overrightarrow{u} _2||\overrightarrow{v} _2||\overrightarrow{g} _2, \cdots ,\overrightarrow{uv} _N||\overrightarrow{v} _N||\overrightarrow{g} _N} \right\}\) by concatenating X, Y and Z, where || is a concatenation operation.
Experiment settings
Multitask combination settings
We design various multitask combinations to answer two key questions.
-
Can joint training of two tasks with great performance (like ‘Alliance between Giants’) achieve higher performance than random combination of two tasks?
The results of single-task-driven SSL suggest that PairDistance, PathClass, and SimCon achieve the relatively higher performance. We thus first chose all combinations of ‘Alliance between Giants’ (that is, PairDistance–PathClass, PathClass–SimCon and PairDistance–SimCon). We next randomly select eight other two-task combinations, that is, EdgeMask–PairDistance, ClusterPre–PathClass, ClusterPre–PairDistance, EdgeMask–PathClass, EdgeMask–SimReg, PairDistance–SimReg, ClusterPre–EdgeMask, SimReg–SimCon.
-
Can the combinations integrating multimodal information further improve the prediction performance?
Based on eleven two-task combinations, we select four multitask combinations to evaluate the influence of different modalities. As shown in Supplementary Table 29, there is only one different task in context compositions. For example, PairDistance–SimCon is turned into PairDistance–EdgeMask–SimCon by adding SimReg. In addition, the pool of combination strategies keep diversity criterions, that is, each task is combined at least five times. We therefore select fifteen kinds of task combinations to guaranteed reliability.
Drug discovery predictions under different scenarios
In this study we focus on the performance of various SSL tasks on DDI and DTI predictions, as they are key stages and play important roles in various applications of drug discovery. Simultaneously, DDI and DTI predictions are treated as link predictions in homogeneous and heterogeneous networks, respectively. Therefore, DDI and DTI predictions can systematically demonstrate the performance of various kinds of SSL tasks and combination strategies. According to the guidance of KGE_NFM5, we design the following two experimental scenarios. Warm-start predictions: given a set of drugs and their known DTIs, we aim to predict other potential interactions between these drugs. All the known interactions are positive samples, and an equal number of negative samples are randomly selected from the unknown interactions. The positive and negative samples are split into a training set (90%) and a testing set (10%). In this situation, the training set may include drugs and targets contained in the test set. The same experimental setting as DTI predictions are used for DDI predictions. In this experimental scenarios, we compare the differences among various SSL tasks for DDI and DTI predictions, and draw a conclusion on which combination strategies can generate the best performance. Cold start for drugs: in real drug discovery, it is more important and challenging to predict potential targets and drugs that may interact with newly discovered chemical compounds. In other words, the test set contains drugs that are unseen in the training set. To be specific, we randomly select 5% drugs, and then all DTI and DDI pairs associated with these drugs are treated as test set. This scenario aims to validate the conclusions that are found in the warm-start predictions. We use the AUPR and AUROC curves as the evaluation metrics for drug discovery. To reduce the data bias and uncertain disturbance, each model is executed ten times, and the average performance is computed. The hyperparameter selections can be found in Supplementary Section 15.
Data availability
All relevant data including the original network and initialization features can be downloaded from https://github.com/pengsl-lab/MSSL.git. Source data are provided with this paper.
Code availability
The source code can be found at https://github.com/pengsl-lab/MSSL.git. In the GitHub repository, we have provided source code that include the data processing of six SSL pretext tasks, GAT-based multitask representation models, and MLP-based DDI or DTI predictors. We also added a description of how to use program, as well as the license and DOI to the code. The license is GNU General Public License v.3.0 and the doi is https://doi.org/10.5281/zenodo.7650518.
References
Dickson, M. & Gagnon, J. P. Key factors in the rising cost of new drug discovery and development. Nat. Rev. Drug Discov. 3, 417–429 (2004).
Scannell, J., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Shen, W. X. et al. Out-of-the-box deep learning prediction of pharmaceutical properties by broadly learned knowledge-based molecular representations. Nat. Mach. Intell. 3, 334–343 (2021).
Chen, D. et al. Algebraic graph-assisted bidirectional transformers for molecular property prediction. Nat. Commun. 12, 1–9 (2021).
Ye, Q. et al. A unified drug–target interaction prediction framework based on knowledge graph and recommendation system. Nat. Commun. 12, 1–12 (2021).
Luo, Y. et al. A network integration approach for drug–target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8, 1–13 (2017).
Cheng, F. et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 9, 1–12 (2018).
Chu, Y. et al. DTI-CDF: a cascade deep forest model towards the prediction of drug–target interactions based on hybrid features. Brief. Bioinformatics 22, 451–462 (2021).
Chu, Y. et al. DTI-MLCD: predicting drug–target interactions using multi-label learning with community detection method. Brief. Bioinformatics. 22, bbaa205 (2021).
Zheng, S. et al. Predicting drug–protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Liu, R., Wei, L. & Zhang, P. A deep learning framework for drug repurposing via emulating clinical trials on real-world patient data. Nat. Mach. Intell. 3, 68–75 (2021).
Ryu, J., Kim, H. & Lee, S. Deep learning improves prediction of drug–drug and drug–food interactions. Proc. Natl Acad. Sci. USA 115, E4304–E4311 (2018).
Lin, S. et al. MDF-SA-DDI: predicting drug-drug interaction events based on multi-source drug fusion, multi-source feature fusion and transformer self-attention mechanism. Brief. Bioinformatics. 23, bbab421 (2022).
Jiménez-Luna, J., Grisoni, F. & Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2, 573–584 (2020).
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks. In Proc. 13th European Conference on Computer Vision 818–833 (Springer, 2014).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Cheng, F., Kovács, I. A. & Barabási, A. L. Network-based prediction of drug combinations. Nat. Commun. 10, 1–11 (2019).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. In Proc. 4th International Conference on Learning Representations (OpenReview.net, 2017).
Veličković, P. et al. Graph Attention Networks. In Proc. 5th International Conference on Learning Representations (OpenReview.net, 2018).
Hamilton, W. L., Ying, R. & Leskovec, J. Inductive representation learning on large graphs. In Proc. 31st International Conference on Neural Information Processing Systems 1025–1035 (MIT Press, 2017).
Zitnik, M., Agrawal, M. & Leskovec, J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 34, i457–i466 (2018).
Ma, T., **ao, C., Zhou, J. & Wang, F. Drug similarity integration through attentive multi-view graph auto-encoders. In Proc. 27th International Joint Conference on Artificial Intelligence 3477–3483 (Morgan Kaufmann, 2018).
Gysi, D. M. et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl Acad. Sci. USA 118, e2025581118 (2021).
Wang, Z., Zhou, M. & Arnold, C. Toward heterogeneous information fusion: bipartite graph convolutional networks for in silico drug repurposing. Bioinformatics 36, i525–i533 (2020).
He, K., Fan, H., Wu, Y., **e, S. & Girshick, R. Momentum contrast for unsupervised visual representation learning. In Proc. 33rd IEEE/CVF Conference on Computer Vision and Pattern Recognition 9729–9738 (IEEE, 2020).
Grill, J. B. et al. Bootstrap your own latent: a new approach to self-supervised learning. In Proc. 34th Conference on Neural Information Processing Systems Vol. 33, 21271–21284 (MIT Press, 2020).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. Conference of the North American Chapter of the Association for Computational Linguistic 4171–4186 (NAACL, 2019)
Brown, T. B. et al. Language models are few-shot learners. In Proc. 34th Conference on Neural Information Processing Systems Vol. 33, 1877–1901 (MIT Press, 2020).
Pham, T. H., Qiu, Y., Zeng, J., **e, L. & Zhang, P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening. Nat. Mach. Intell. 3, 247–257 (2021).
Wang, Y., Min, Y., Chen, X. & Wu, J. Multi-view graph contrastive representation learning for drug–drug interaction prediction. In Proc. 30th Web Conference 2921–2933 (ACM, 2021).
Wang, X. et al. DeepR2cov: deep representation learning on heterogeneous drug networks to discover anti-inflammatory agents for COVID-19. Brief. Bioinform 22, 1–14 (2021).
Chu, Y. et al. A transformer-based model to predict peptide-HLA class I binding and optimize mutated peptides for vaccine design. Nat. Mach. Intell. 4, 300–311 (2022).
Rong, Y. et al. Self-supervised graph transformer on large-scale molecular data. In Proc. 34th Conference on Neural Information Processing Systems 33, 12559–12571 (MIT Press, 2020).
Wang, X. et al. BioERP: biomedical heterogeneous network-based self-supervised representation learning approach for entity relationship predictions. Bioinformatics 37, 4793–4800 (2021).
Hu, W. et al. Strategies for pre-training graph neural networks. In Proc. 8th International Conference on Learning Representations (OpenReview.net, 2020).
**, W. et al. Self-supervised learning on graphs: deep insights and new direction. Preprint at ar**v https://doi.org/10.48550/ar**v.2006.10141 (2020).
Zeng, X. et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 11, 1775–1797 (2020).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Deng, Y. et al. A multimodal deep learning framework for predicting drug–drug interaction events. Bioinformatics 36, 4316–4322 (2020).
Belkin, M. & Niyogi, P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 15, 1373–1396 (2003).
Ahmed, A., Shervashidze, N., Narayanamurthy, S., Josifovski, V. & Smola, A. J. Distributed large-scale natural graph factorization. In Proc. 22nd International World Wide Web Conference 37–48 (ACM, 2013).
Perozzi, B., Al-Rfou, R. & Skiena, S. Deepwalk: online learning of social representations. In Proc. 20th International Conference on Knowledge Discovery and Data Mining 701–710 (ACM, 2014).
Liu, B. & Tsoumakas, G. Optimizing area under the curve measures via matrix factorization for predicting drug–target interaction with multiple similarities. Preprint at ar**v https://arxiv.org/abs/2105.01545 (2021).
Wang, Y., Min, Y., Chen, X. & Wu, J. Multi-view graph contrastive representation learning for drug–drug interaction prediction. In Proc. 30th Web Conference 2921–2933 (ACM, 2021).
Hou, W. & Cronin, S. B. A review of surface plasmon resonance-enhanced photocatalysis. Adv. Funct. Mater. 23, 1612–1619 (2013).
Sigman, M. & Cecchi, G. A. Global organization of the Wordnet lexicon. Proc. Natl Acad. Sci. USA 99, 1742–1747 (2002).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Costa, L. D. F., Rodrigues, F. A., Travieso, G. & Villas Boas, P. R. Characterization of complex networks: a survey of measurements. Adv. Phys. 56, 167–242 (2007).
Peng, Z., Dong, Y., Luo, M., Wu, X. M. & Zheng, Q. Self-supervised graph representation learning via global context prediction. Preprint at ar**v https://arxiv.org/abs/2003.01604 (2020).
Fu, G. et al. Predicting drug target interactions using meta-path-based semantic network analysis. BMC Bioinf. 17, 1–10 (2016).
Wu, G., Liu, J. & Yue, X. Prediction of drug–disease associations based on ensemble meta paths and singular value decomposition. BMC Bioinf. 20, 1–13 (2019).
Breiman, L. Random Forests. Mach. Learn. 45, 5–32 (2001).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Yang, X., Deng, C., Dang, Z., Wei, K. & Yan, J. SelfSAGCN: self-supervised semantic alignment for graph convolution network. In Proc. 34th IEEE/CVF Conference on Computer Vision and Pattern Recognition 16775–16784 (IEEE, 2021).
Kapidis, G., Poppe, R. & Veltkamp, R. C. Multi-dataset, multitask learning of egocentric vision tasks. IEEE Trans Pattern Anal. Mach. Intell. https://doi.org/10.1109/TPAMI.2021.3061479 (2021).
Hernández-Lobato, D. & Hernández-Lobato, J. M. Learning feature selection dependencies in multi-task learning. In Proc. 27th Conference on Neural Information Processing Systems 746–754 (MIT Press, 2013).
Zhao, S., Liu, T., Zhao, S. & Wang, F. A neural multi-task learning framework to jointly model medical named entity recognition and normalization. In Proc. 33rd AAAI Conference on Artificial Intelligence 817–824 (AAAI, 2019).
Baxter, J. A Bayesian/information theoretic model of learning to learn via multiple task sampling. Mach. Learn. 28, 7–39 (1997).
Ruder, S. An overview of multi-task learning in deep neural networks. Preprint at ar**v https://doi.org/10.48550/ar**v.1706.05098 (2017).
Wang, Y., Wang, J., Cao, Z. & Barati Farimani, A. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4, 279–287 (2022).
Zeng, J. & **e, P. Contrastive self-supervised learning for graph classification. In Proc. 35th AAAI Conference on Artificial Intelligence 10824–10832 (AAAI, 2021).
Li, T., Wang, L. & Wu, G. Self-supervision to distillation for long-tailed visual recognition. In Proc. 34th IEEE/CVF International Conference on Computer Vision 630–639 (IEEE, 2021).
Li, Y, et al. GMSS: graph-based multi-task self-supervised learning for EEG emotion recognition. IEEE Trans. Affect. Comput. https://doi.org/10.1109/TAFFC.2022.3170428 (2022).
Law, V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–D1097 (2014).
Hernandez, T. et al. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic Acids Res. 36, D913–D918 (2008).
Zhu, F. et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 40, D1128–D1136 (2012).
Keshava Prasad, T. et al. Human protein reference database 2009 update. Nucleic Acids Res. 37, D767–D772 (2009).
Figeys, D. Map** the human protein interactome. Cell Res. 18, 716–724 (2008).
Oughtred, R. et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019).
Brown, A. S. & Patel, C. J. A standard database for drug repositioning. Sci. Data 4, 170029 (2017).
Ursu, O. et al. DrugCentral: online drug compendium. Nucleic Acids Res. 45, D932–D939 (2017).
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A. & McKusick, V. A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005).
Davis, A. P. et al. The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 41, D1104–D1114 (2013).
Bodenreider, O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Watts, D. J. Networks, dynamics, and the small-world phenomenon. Am. J. Sociol. 105, 493–527 (1999).
Taylor, W. L. ‘Cloze procedure’: a new tool for measuring readability. Journalism Quarterly 30, 415–433 (1953).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and eccoding rules. J. Chem. Inf. Model. 28, 31–36 (1988).
Vilar, S. et al. Similarity-based modeling in large-scale prediction of drug–drug interactions. Nat. Protoc. 9, 2147–2163 (2014).
Gotoh, O. An improved algorithm for matching biological sequences. J. Mol. Biol. 162, 705–708 (1982).
Ni, P. et al. Constructing disease similarity networks based on disease module theory. IEEE/ACM Trans. Comput. Biol. Bioinform. 17, 906–915 (2018).
Liu, P., Qiu, X. & Huang, X. Adversarial multi-task learning for text classification. In Proc. 55th Annual Meeting of the Association for Computational Linguistics 1–10 (ACL, 2017).
Bousmalis, K., Trigeorgis, G., Silberman, N., Krishnan, D. & Erhan, D. Domain separation networks. Proc. 30th Conference on Neural Information Processing Systems Vol. 29, 343–351 (2016).
Collobert, R. & Weston, J. A unified architecture for natural language processing: deep neural networks with multitask learning. In Proc. 25th International Conference on Machine Learning 160–167 (ACM, 2008).
Acknowledgements
This work was supported by the NSFC (grant nos. U19A2067 and 81973244), the National Key R&D Program of China (grant no. 2022YFC3400400), The Funds of Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDB38040100), The Funds of State Key Laboratory of Chemo/Biosensing and Chemometrics, and the National Supercomputing Center in Changsha and Peng Cheng Laboratory.
Author information
Authors and Affiliations
Contributions
X.W. and Y.C. conceived the original idea and developed the code for the core algorithm. S.P designed the experiment and wrote the initial version of the manuscript. F.L. and Y.Y analysed the experimental data and edited this manuscript. Y.N.Y constructed the biomedical network data. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks ShuaiCheng Li, Dong-qing Wei and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
The Supplementary information file includes important experiment results and method description.
Supplementary Data
This compressed file contains source data of Supplementary Figs. 1–10: (1) Source_Data_Supplementary_FigS1.xlsx, the results of self-supervised tasks for cold-start predictions; (2) Source_Data_Supplementary_FigS2.xlsx, heatmap of two-task combinations for cold-start predictions where the results are normalized to [0,1] along the x-axis via the min–max normalization technique; (3) Source_Data_Supplementary_FigS3.xlsx, the results obtained with multimodal SSL tasks for cold-start drug discovery; (4) Source_Data_Supplementary_FigS4.xlsx, heatmap of two-task combinations on Luo’s dataset for warm-start predictions where the results are normalized to [0,1] along the x-axis via min–max normalization technique; (5) Source_Data_Supplementary_FigS5.xlsx, the results obtained with multimodal SSL tasks on Luo’s dataset for warm-start drug discovery; (6) Source_Data_Supplementary_FigS6.xlsx, performance of all methods on different splitting ratios; (7) Source_Data_Supplementary_FigS8.xlsx, root-mean-square deviation of the protein in two complex system as the 100 function of simulation time; (8) Source_Data_Supplementary_FigS10.xlsx, surface plasmon resonance sensorgrams and binding affinity between two molecules and IL-6; (9) Source_Data_Supplementary_FigS13.xlsx, performance comparison of PESReM and PairDistance–EdgeMask–SimCon for DTI predictions where PESReM denotes the variant of PairDistance–EdgeMask–SimCon when test data is removed from SSL; and (10) Source_Data_Supplementary_FigS14.xlsx, the distribution of node degree.
Source data
Source Data Fig. 3
The results of self-supervised tasks for drug warm-start predictions.
Source Data Fig. 4
Heatmap of two-task combinations for drug warm-start predictions.
Source Data Fig. 5
The results obtained multimodal task combinations for drug warm-start predictions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Cheng, Y., Yang, Y. et al. Multitask joint strategies of self-supervised representation learning on biomedical networks for drug discovery. Nat Mach Intell 5, 445–456 (2023). https://doi.org/10.1038/s42256-023-00640-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00640-6
- Springer Nature Limited
This article is cited by
-
HoopTransformer: Advancing NBA Offensive Play Recognition with Self-Supervised Learning from Player Trajectories
Sports Medicine (2024)
-
Self-supervised learning for medical image analysis: a comprehensive review
Evolving Systems (2024)