Abstract
The systematics of the Cricetid genus Neodon have long been fraught with uncertainty due to sampling issues and a lack of comprehensive datasets. To gain better insights into the phylogeny and evolution of Neodon, we systematically sampled Neodon across the Hengduan and Himalayan Mountains, which cover most of its range in China. Analyses of skulls, teeth, and bacular structures revealed 15 distinct patterns corresponding to 15 species of Neodon. In addition to morphological analyses, we generated a high-quality reference genome for the mountain vole and generated whole-genome sequencing data for 47 samples. Phylogenomic analyses supported the recognition of six new species, revealing a long-term underestimation of Neodon diversity. We further identified positively selected genes potentially related to high-elevation adaptation. Together, our results illuminate how climate change caused the plateau to become the centre of Neodon origin and diversification and how mountain voles have adapted to the hypoxic high-altitude plateau environment.
Similar content being viewed by others
Introduction
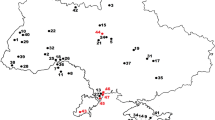
Voles constitute one of the youngest groups of rodents. Mountain voles, belonging to genus Neodon (Rodentia: Cricetidae), which occur only in the Tibetan-Himalayan region (THR) (Fig. 1)26, along with 1X–15X whole-genome sequencing (WGS) data for each morphologically distinct taxon (Supplementary Data 2). This extensive dataset allowed the identification and description of six new species of Neodon. Our analyses showed that rapid climate change, complex topography and founder events resulting from dispersal were the key factors driving Neodon diversification and evolution. In addition, our de novo genome assembly revealed the genetic basis of the adaptations of mountain voles to high-latitude environments, characterised by pressures such as hypoxia, high UV radiation and low temperatures.
Results
Morphological evidence of six unidentified lineages of Neodon
We analysed 235 specimens of Neodon to examine the possible existence of new species of Neodon based on morphological evidence. We also generated WGS data for 48 specimens to explore their genetic divergence and potential taxonomic status (Supplementary Data 1 and 2).
Initial observations of skulls, teeth and bacular structures showed 15 distinct patterns (Fig. 2 and Supplementary Fig. 1), each representing a putative species of Neodon. This included eight described species1,24, one previously evaluated group with unclear status and six tentatively unidentified taxa. We recorded the characteristics of the genitalia and bacular structures for males; and dental, external and cranial measurements for both sexes (detailed abbreviations of the measurements are provided in Supplementary Data 3) and further conducted morphological comparisons of these 15 putative species. The morphology of the glans penis provided useful clues about the affinities of microtine species and the differences in the characters of the glans penis clearly distinguished all putative species (Fig. 2). The pairwise Euclidean distances of dental measurements (e.g., the number of closed triangles on the first lower molar) also distinguished 15 patterns (Supplementary Fig. 2 and Supplementary Data 4). In addition, principal component analysis (PCA) (Supplementary Fig. 3) and subsequent two-sided t-tests or Wilcoxon rank-sum tests (Supplementary Fig. 4) of 17 statistical measurements of external and cranial characteristics of 95 intact adults (Supplementary Data 5–7) also resolved all 15 putative species.
a Comparison of tooth rows. b Comparison of glans penes. Numbered views are 1: glans; 2: midventral cut view; 3: urethral lappet; 4: dorsal papilla. Lettered structural features in a1 and a2 are a. distal baculum; b. outer crater; c. inner crater; d. ventral groove; e. glans; f. prepuce; g. penis body; h. station of dorsal papilla; i. lateral baculum (cartilage); j. urethral lappet; k. lateral baculum (bony part); l. distal baculum (bony part); and m. proximal baculum. The taxa are (from top to bottom) Neodon leucurus, N. fuscus, N. linzhiensis, N. forresti, N. irene from Clade 1, N. nyalamensis, N. sikimensis, unidentified taxon 1 (from Nanyi township, Milin County), unidentified taxon 2 (from Shergyla Mountains, Linzhi county), unidentified taxon 3 (from Motuo County, south of the Namchabarwa Mountains) from Clade 2, N. medogensis, unidentified taxon 4 (from Ridong village, Bershula Mountains, Chayu County), N. clarkei, unidentified taxon 5 (from Bomi County) and unidentified taxon 6 (from Chibagou National Nature Reserve, Chayu County) from Clade 3 (refer to Fig. 3 for clade information).
Molecular evidence for all lineages of Neodon
We generated a total of 241 Gb of 10X Genomics linked-reads (67.56X) for one specimen of Neodon sampled from north of the Yarlung Zangbo River on Shergyla Mountain (unidentified taxon 2), and produced a genome assembly with a total length of 2.25 Gb and a scaffold N50 of 10.85 Mb. In addition, we obtained a total of 620.45 Gb of reads for an additional 47 samples, with the amount of data generated from each sample ranging from 2.21 Gb to 43.38 Gb (Supplementary Data 2). A total of 4951 full-length single-copy orthologous gene groups were annotated in our reference genome using BUSCO27. The “nuclear gene set” obtained for all lineages after the removal of low-confidence genes, consisted of a total of 4624 coding genes, with an average, maximal and minimal lengths of 1885 nt, 23,046 nt and 222 nt, respectively (Supplementary Fig. 5).
We obtained complete mitogenomes for sequenced specimens and calculated Kimura 2-parameter genetic distances for mitochondrial protein-coding genes (Supplementary Fig. 6 and Supplementary Data 8–10). The results for cox1 and cytb, the two most-widely used barcoding genes in mammals, showed an average interspecific genetic distances of 11.00% for cox1 and 11.30% for cytb. Species delimitation methods (bayesian implementation of Poisson Tree Processes (bPTP), automatic barcode gap discovery (ABGD) and BPP) based on mitochondrial or nuclear datasets recognised the same 15 species, including all six undescribed morphological lineages (Supplementary Figs. 7–10). Furthermore, the species delimitation results identified a split within N. sikimensis (Supplementary Fig. 7), and these specimens showed the greatest intraspecific genetic distances (average of 5.23% for cox1 and 5.20% for cytb). However, the specimens of N. sikimensis, which were collected in the same region at the same time, did not differ significantly in their morphology. This likely indicated the cooccurrence of divergent mitogenomes, as reported in the Asian elephant28 and other species. Further work can explore the possibility of unabated gene flow (one species) or restricted gene flow (two species).
Phylogenetic analysis corroborates taxonomic status
The analyses generated eight phylogenetic trees, among which two were based on barcoding genes (Supplementary Fig. 11), two were based on 13 mitochondrial coding genes (Supplementary Fig. 7), and four were based on 4,624 nuclear genes (Supplementary Fig. 10). All nuclear trees generated by coalescent and concatenation approaches shared the same topology, with only small-scale incongruences being identified between the nuclear trees and the other four mitochondrial gene trees. We used the nuclear ASTRAL III tree as the species tree in downstream analyses (Supplementary Fig. 10a). The phylogenetic analysis recovered Neodon as sister to Lasiopodomys, sharing a common ancestor with Microtus and Alexandromys, and the species of Neodon formed three major clades.
Description of six new species
Multiple resources supported the recognition of six new morphological species. Thus we described these species as follows (Expanded description in Supplementary Note 1, 2). All type series specimens have been deposited with the Sichuan Academy of Forestry:
Neodon namchabarwaensis Liu SY., Zhou CR., Murphy WR. & Liu SL., sp. nov. (unidentified taxon 1)
Holotype
Adult female, field number XZGB0818009, collected by Liao Rui on 10 August 2008. Specimen preserved as a skin, cleaned skull, and tissues. Skull, dentition and mandible in Supplementary Fig. 1a.
Type locality
Nanyi township, Milin County, south of **zang, China, 29.17889° E, 94.15113° N, elevation 3160 m a.s.l.
Paratypes
Five specimens topotypes (3♂♂, 2♀♀), field numbers: XZGB0817007♂, XZGB0818006♀, XZGB0818010♂, XZGB0828001♀, XZGB09N235♂;
Distribution
Known from south of the Yarlung Zangbo River, north of the Namchabarwa Mountains. The lowest elevation is 3130 m a.s.l.
Etymology
Species named for the famous Namcha Barwa Mountain, the highest mountain in this region where the new species occurs.
Diagnosis
Medium body, average length 114.9 mm (adult); average hind foot length 20.1 mm. Average tail length 46.4 mm, approximately 40.4% of HBL. First lower molar with 3 closed triangles in front of the posterior transverse space, 6 inner and 5 outer angles. 1st upper molar with 4 inner and 3 outer angles. 2nd upper molar with 3 inner and 3 outer angles. 3rd upper molar with 4 inner and 3 outer angles.
Neodon shergylaensis Liu SY., Zhou CR., Murphy WR. & Liu SL., sp. nov. (unidentified taxon 2)
Holotype
Adult male, field number XZGB09N195, collected by Liao Rui and Liu Yang on 30 May 2009. Specimen preserved as a skin, cleaned skull, penis and tissues. Skull, dentition, and mandible are in Supplementary Fig. 1b.
Type locality
Shergyla Mountains of Linzhi county, southeast of **zang, China, 29.62368° E, 94.66174° N, elevation 4500 m.
Paratypes
6 specimens (1♂, 5♀♀), field numbers: LZRAP01013♂, LZRAP01020♀, LZRAP01014♀, LZRAP01019♀, XZGB09N197♀, GB0815001J♀
Distribution
Known from north of the Yarlung Zangbo River at over 3160 m a.s.l. both sides of Shergyla Mountains and Niyang River.
Etymology
The species is named for its type locality. This region supports a high biodiversity.
Diagnosis
Medium body, average length 115.7 mm (adult); average hind foot length 19.5 mm. Average tail length 42.7 mm, approximately 37% of HBL. The first lower molar with 3 closed triangles in front of the posterior transverse space, 6 inner and 4 outer angles in 63% specimens, 6 inner and 5 outer angles in 37% specimens. 1st upper molar with 3 inner and 3 outer angles. 2nd upper molar with 3 inner and 3 outer angles. 3rd upper molar with 4 inner and 3 outer angles.
Neodon liaoruii Liu SY., Zhou CR., Meng GL. & Liu SL., sp. nov. (unidentified taxon 3)
Holotype
Adult male, field number XZ11117, collected by Liao Rui on 1 November 2011. Specimen preserved as a skin, cleaned skull, penis and tissues. Skull, dentition and mandible in Supplementary Fig. 1c.
Type locality
Motuo County, south of **zang, China, 29.47028° E, 94.984° N, elevation 3260 m.
Paratypes
Ten specimens (4♂♂, 6♀♀), field numbers: MT11036♀, MT11066♀, MT11067♀, MT11109♂, MT11118♂, MT11120♂, MT11122♀, MT11142♂, MT11143♀, MT11144♀.
Distribution
Known from south of the Namchabarwa Mountains. Lowest elevation 2660 m a.s.l.
Etymology
Species epithet is a patronym for the collector, Mr. Liao Rui. He made an important contribution to our collecting specimens.
Diagnosis
Relatively large body, average length 116.8 mm (adult); average hind foot length 21.1 mm. Avarage tail length 59.3 mm, ~50.8% of HBL. First lower molar with 3 closed triangles in front of the posterior transverse space, 6 inner and 5 outer angles. 1st upper molar with 3 inner and 3 outer angles. 2nd upper molar with 2 inner and 3 outer angles in 66% specimens, and 3 inner and 3 outer angles in another 34% specimens. 3rd upper molar with 4 inner and 3 outer angles in 61% specimens, and 3 inner and 3 outer angles in another 39% specimens.
Neodon bershulaensis Liu SY., Zhou CR., Liu Y. & Liu SL., sp. nov. (unidentified taxon 4)
Holotype
Adult male, field number XZ11010, collected by Liao Rui on 3 Mach 2011. Specimen preserved as a skin, cleaned skull, penis and tissues. Skull, dentition, and mandible in Supplementary Fig. 1d.
Type locality
Ridong village, Bershula Mountains, Chayu County, southeast of **zang, China. 98.12407° E, 28.58392° N, elevation 3750 m a.s.l.
Paratypes
3 intact adult specimens, field number: CHYRD-03♀, CHYRD-04♂, CSD3825♂.
Distribution
Known from the type locality only, Ridong village, Chayu County, southeast of **zang.
Etymology
Species epithet for the famous Bershula Mountains, where type locality, Ridong is at its foot.
Diagnosis
Medium body, average length 107 mm (adult); hind feet length 18–20 mm (average 19 mm). Average tail length 51.5 mm, 48.1% of HBL. First lower molar with 5 closed triangles in front of the posterior transverse space, 6 inner and 4 outer angles. 1st upper molar with 4 inner and 3 outer angles in 70% specimens; other 30% with 3 inner and 3 outer angles. 2nd upper molar with 3 inner and 3 outer angles. 3rd upper molar with 4 inner and 3 outer angles.
Neodon bomiensis Liu SY., Zhou CR., Meng GL. & Liu SL., sp. nov. (unidentified taxon 5)
Holotype
Adult male, field number XZ13015, collected by Liao Rui on 31 October 2013. Specimen preserved as a skin, cleaned skull, penis and tissues. Skull, dentition and mandible in Supplementary Fig. 1e.
Type locality
Bomi County, southeast of **zang, China, 95.9575816° E, 29.82959° N, elevation 2900 m a.s.l.
Paratypes
2 intact adults specimens, field numbers: MT11304♀, XZ13016♀
Distribution
Known only from the type locality, Bomi County, southeast of **zang.
Etymology
Species epithet derived from the county where type series collected.
Diagnosis
Medium body, average length 111.75 mm (adult); hind feet length 18–19 mm (average 18.75 mm). Tail length 53–56 mm (average 53.75 mm), approximately 48.1% of HBL. First lower molar with 4 closed triangles in front of the posterior transverse space, 6 inner and 4 outer angles in 60% specimens; other 40% with 5 inner and 4 outer angles. 1st upper molar with 4 closed triangles after the anterior transverse space, forming 3 inner and 3 outer angles. 2nd upper molar with 3 inner and 3 outer angles, and the last inner angle much small. 3rd upper molar with 3 inner and 3 outer angles.
Neodon chayuensis Liu SY., Zhou CR., Liu Y., Tang MK. & Liu SL., sp. nov. (unidentified taxon 6)
Holotype
Adult female, field number CY37, collected by Liu Yang on 8 October 2007. Specimen preserved as a skin, cleaned skull and tissues. Skull, dentition, and mandible in Supplementary Fig. 1f.
Type locality
Chibagou National Nature Reserve, Chayu County, southeast of ** more than 50% with the adaptor sequence, with a maximal 1 bp mismatches to the adaptor sequence); or (3) more than 30% of the read length below Q30 (Supplementary Data 2).
A total of 1,612.36 million paired-dnd reads were generated with 10X technology was generated for Neodon shergylaensis sp. nov., and the reference genome was assembled using SuperNova v2.1.158, with a preset genome size of 2.50 Gb and a weighted mean molecule size of 18.39 kb. One of the two pseudohaplotypes generated using SuperNova with “--maxreads = ‘all’ --accept-extreme-coverage, --style = pseudohap2” was used to obtain the core gene set using BUSCO v3.0.127. In addition, the assembled genome was annotated using MAKER2 v2.31.1059,60 (control files can be found in Supplementary Data 16) for further evolutionary analysis. We also assembled and annotated the mitogenome of each sample using MitoZ61 with ~3 Gb (~1X) of filtered data61.
Gene dataset construction
A read-map**-then-consensus-calling pipeline was used to obtain orthologous nuclear genes for each sample. For this purpose, (1) we obtained complete and single-copy orthologues of the N. shergylaensis sp. nov. genome using BUSCO v3.0.127 with a database of the Euarchontoglires group (6192 genes, v2), and we removed orthologues with high homologue to each other (BLASTn v2.6.0+ with e value < 1e-5)62 to avoid map** uncertainty in the subsequent steps, deleted orthologues with internal stop codons, extracted corresponding genomic regions for the remaining qualified orthologues with custom Python scripts and then used the residual data as a reference for the resequenced samples. (2) We mapped the WGS data of each sample onto the reference using BWA-MEM v0.7.171 (Supplementary Fig. 17 and Supplementary Data 18 and 19). The “map**-derived” gene dataset was used in subsequent analyses.
Species delimitation
We calculated the Kimura 2-parameter genetic distances between lineages for each gene using the dist.dna function in R ape v1.1-168 and explored the correlation between genetic distance and geographic distance. Geographic distances between different sampling sites were calculated with the geopy.distance.geodesic function in Python geopy v2.0.0 package69 (https://github.com/geopy/geopy). Pearson correlation coefficients were calculated with the cor function in R and plotted using ggpubr70. In addition to morphological identification, we conducted species delimitation analysis using the clustering-based method bPTP71 based on both the mitochondrial and nuclear trees. We also applied the similarity-based ABGD72 and the multispecies coalescent (MSC) model-based BPP v4.3.873 analysis using only the mitochondrial data. We also applied both the A1074,75 and A1176 analyses implemented in BPP to the 31 Neodon specimens using mitochondrial data. Specimens of each morphological species were grouped into the same population, including 1 to 5 specimens of each morphological species. Species delimitation were performed with a user-specified guide tree in the A10 analysis, while species delimitation and species tree inference were jointly calculated in the A11 analysis (detailed in Supplementary Note 1).
Phylogenetic inference
Both coalescent and concatenation methods were used to infer phylogenetic trees. We inferred the best maximum likelihood trees using RAxML v8.2.1277 with the GTR + GAMMA model from 20 independent tree searches and 500 bootstrap replicates for each gene, and then obtained the final species tree using ASTRAL-III78 based on the multispecies coalescent model with the bootstrap support of each node being estimated by the multilocus resampling method79. SVDquartets (parameters of “eval Quartets = 1e + 6 bootstrap = standard”) implemented in PAUP v4.0a16780,81 was also utilised to estimate the species tree with the same dataset to validate the results. Additionally, we concatenated the gene alignments to generate a “supergene” alignment for each species and obtained species trees using IQ-tree v1.6.1282, RAxML or MrBayes83. The inferred phylogenetic tree comprised two gene sets - the “mitochondrial Gene Set” and the “nuclear Gene Set”. All trees inferred from the nuclear dataset showed the same topology, and we thus re-estimated the branch lengths of the final species tree in units of substitutions per site using ExaML v3.0.2184.
Divergence time estimation
We estimated the divergence times of lineages of Neodon based on the second codon sites of nuclear genes using MCMCTree, a Bayesian relaxed clock method implemented in the PAML v4.9 h package30. For estimation, an approximate likelihood calculation of the ‘REV’ (GTR, model = 7) model was applied, and multiple fossil calibration points taken from records in the Palaeobiology Database (Accessed 2018 Dec 12)85 and the timetree database86 were included, as follows: (a) the root age was set as 7.9 Mya, as supported by the occurrence of Promimomys in the fossil record;87 (b) the splits of Lasiopodomys and Neodon, Eothenomys and Myodes were calibrated as <0.53 Mya based on the earliest occurrence of the oldest fossil record of Myodes from the Paleobiology Database and another fossil record of Promimomys;87 (c) the split data of Eothenomys was set as 2.7–5.3 Mya based on the fossil calibration point of Eothenomys (3.6–2.6 Mya);88,89 and (d) the split data of Microtus and Alexandromys was dated between 0.6 and 3.5 Mya based on previous studies90,91. The minimum boundary was supported by the earliest occurrence of Allophaiomys in the fossil record in the database. BaseML first estimated a prior substitution rate, and MCMCTree then generated the Gradient and Hessian matrices with following settings: ‘correlated rates clock’ (clock = 3), overall substitution rate (rgene gamma) set of G (1, 12.0), and rate drift parameter (sigma2 gamma) set of G (1, 4.5). Next, we conducted two independent MCMC runs with different random seed numbers and a burn-in of 500,000 iterations to check for convergence. Each run was sampled every 1000 iterations until 500,000 samples had accumulated. We also applied Tracer v1.7.197 was applied to perform KEGG and GO enrichment annotation. Protein structure was predicted using SWISS-MODEL98.
Statistics and reproducibility
The morphometric variation in non-sex-related measurements of adult specimens was analysed using PCA in SPSS v17.0. We employed Kaiser-Meyer-Olkin and Bartlett’s tests to check the fitness of the PCA, followed by Tukey’s test. Independent-samples two-sided t tests or Wilcoxon rank-sum tests were also performed to check the differences between the taxon pairs after PCA. The significant positively selected genes were confirmed using Bonferroni test. Reproducibility was confirmed by performing analyses with independent replicates (for morphological analyses), five hundred bootstrap replicates or different coalescent and concatenation approaches as described in the Methods section.
Nomenclatural Acts
This published work and the nomenclatural acts it contains have been registered in ZooBank, the proposed online registration system for the International Code of Zoological Nomenclature (ICZN). The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix “http://zoobank.org/”. The LSID for this publication is: urn:lsid:zoobank.org:pub:794808AA-EA46-4E86-B482-9983214688BB.
The LSID for Neodon namchabarwaensis Liu SY., Zhou CR., Murphy WR. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:8B19E76E-2E5F-452E-A94B-0824DB45CB30
The LSID for Neodon shergylaensis Liu SY., Zhou CR., Murphy WR. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:811C522A-2B13-48EE-A8B4-3B758E3EB129
The LSID for Neodon liaoruii Liu SY., Zhou CR., Meng GL. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:D4E07979-F92F-4825-BA6B-BB5B6881F9FD
The LSID for Neodon bershulaensis Liu SY., Zhou CR., Liu Y. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:A72C6927-1269-4E65-8183-6F48D86F06E9
The LSID for Neodon bomiensis Liu SY., Zhou CR., Meng GL. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:445E9955-1D43-41E9-AE51-71A3CDFDB28D
The LSID for Neodon chayuensis Liu SY., Zhou CR., Liu Y., Tang MK. & Liu SL., sp. nov. is: urn:lsid:zoobank.org:act:0F26DDC2-C279-4DE6-AAF2-B1E4C9917B6F
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data that support our findings have been deposited in the NCBI database under BioProject PRJNA564473 (ncbi.nlm.nih.gov/bioproject/?term= PRJNA564473) and CNGB Nucleotide Sequence Archive (CNSA) under the accession number CNP0000173 (https://db.cngb.org/search/project/CNP0000173). Source data for figures can be found in Supplementary Data 20.
Code availability
All analyses were performed using open source software tools, and the detailed parameters for each tool are shown in the relevant methods. Custom scripts are archived at GitHub (https://github.com/linzhi2013/busco_process_scripts).
References
Liu, S. et al. Taxonomic position of Chinese voles of the tribe Arvicolini and the description of 2 new species from **zang, China. J. Mammal. 98, 166–182 (2017).
Lv, X., **a, L., Ge, D., Wu, Y. & Yang, Q. Climatic niche conservatism and ecological opportunity in the explosive radiation of arvicoline rodents (Arvicolinae, Cricetidae). Evolution 70, 1094–1104 (2016).
Abramson, N., Lebedev, V., Bannikova, A. & Tesakov, A. Radiation events in the subfamily Arvicolinae (Rodentia): Evidence from nuclear genes. In Doklady Biological Sciences. 458–461 (Springer, 2009).
Mosbrugger, V., Favre, A., Muellner-Riehl, A. N., Päckert, M. & Mulch, A. Cenozoic evolution of geo-biodiversity in the Tibeto-Himalayan region. In Mountains, Climate, and Biodiversity.429–449 (Wiley-Blackwell, 2018).
Muellner-Riehl, A. N. Mountains as evolutionary arenas: patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto-Himalayan region. Front. Plant Sci. 10, 195 (2019).
**ng, Y. & Ree, R. H. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl Acad. Sci. USA 114, E3444–E3451 (2017).
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A. & Kent, J. Biodiversity hotspots for conservation priorities. Nature 403, 853 (2000).
Marchese, C. Biodiversity hotspots: a shortcut for a more complicated concept. Glob. Ecol. Conserv. 3, 297–309 (2015).
Tamma, K. & Ramakrishnan, U. Higher speciation and lower extinction rates influence mammal diversity gradients in Asia. BMC Evol. Biol. 15, 11 (2015).
Moreau, C. S. & Bell, C. D. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution 67, 2240–2257 (2013).
Pisano, J. et al. Out of Himalaya: the impact of past Asian environmental changes on the evolutionary and biogeographical history of Dipodoidea (Rodentia). J. Biogeogr. 42, 856–870 (2015).
Weigold, H. Die Biogeographie Tibets Und Seiner Vorländer (Verein Sächsischer Ornithologen, 2005).
Jia, D. R. et al. Out of the Qinghai–Tibet Plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophaë rhamnoides (Elaeagnaceae). New Phytol. 194, 1123–1133 (2012).
Wang, X., Tseng, Z. J., Li, Q., Takeuchi, G. T. & **e, G. From ‘third pole’to north pole: a Himalayan origin for the arctic fox. Proc. R. Soc. B Biol. Sci. 281, 20140893 (2014).
Wang, X. et al. Out of Tibet: genomic perspectives on the evolutionary history of extant pikas. Mol. Biol. Evol. 37, 1577–1592 (2020).
Hodgson, B. H. Classified catalogue of mammals of Nepal, (corrected to end of 1841, first printed in 1832). J. Asiatic Soc. Bengal 10, 907–916 (1841).
Allen, G. M. Mammals of China and Mongolia. Part 2. Central Asiatic Expeditions 1921-1930. Vol. 11, p. 621–1350 (American Museum of Natural History, 1940).
Carleton, M. D. & Musser, G. G. Systematic studies of oryzomyine rodents (Muridae, Sigmodontinae)-definition and distribution of Oligoryzomys vegetus (Bangs, 1902). Proceedings of the Biological Society of Washington (Biological Society of Washington, 1995).
Gromov, I. & Polyakov, I. Y. Mammals: voles (microtinae). Fauna SSSR 3, 1–504 (1977).
Corbet, G. B. The mammals of the Palaearctic region: a taxonomic review. British Museum (Natural History) (1978).
Ellerman, J. R. & Morrison-Scott, T. C. S. Checklist of Palaearctic and Indian Mammals, 1758–1946. (British Museum of Natural History, 1951).
Ellerman, J. R. The Families and Genera of Living Rodents, Volume III, Part I. (British Museum of Natural History, London, 1949).
Musser, G. G. & Carleton, M. D. Order rodentia. Mammal Species of the World: A Taxonomic and Geographic Reference 2 (2005).
Liu, S. et al. A new vole from **zang, China and the molecular phylogeny of the genus Neodon (Cricetidae: Arvicolinae). Zootaxa 3235, 1–22 (2012).
Pradhan, N. et al. Further assessment of the Genus Neodon and the description of a new species from Nepal. PLoS ONE 14, e0219157 (2019).
Zheng, G. X. Y. et al. Haploty** germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34, 8 (2016).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Vidya, T., Sukumar, R. & Melnick, D. J. Range-wide mtDNA phylogeography yields insights into the origins of Asian elephants. Proc. R. Soc. B: Biol. Sci. 276, 893–902 (2009).
Storz, J. F., Scott, G. R. & Cheviron, Z. A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 213, 4125–4136 (2010).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Nguyen, H. D. et al. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847.e834 (2017).
Smith, E. A. et al. DEK is required for homologous recombination repair of DNA breaks. Sci. Rep. 7, 1–12 (2017).
Sousounis, K. et al. Eya2 promotes cell cycle progression by regulating DNA damage response during vertebrate limb regeneration. Elife 9, e51217 (2020).
Elkholi, R. et al. MDM2 integrates cellular respiration and apoptotic signaling through NDUFS1 and the mitochondrial network. Mol. Cell 74, 452–465.e457 (2019).
Wikramanayake, T. C., Nicu, C., Chéret, J., Czyzyk, T. A. & Paus, R. Mitochondrially localized MPZL3 emerges as a signaling hub of mammalian physiology. BioEssays 43, 2100126 (2021).
Kim, Y. H. et al. A MST1–FOXO1 cascade establishes endothelial tip cell polarity and facilitates sprouting angiogenesis. Nat. Commun. 10, 1–17 (2019).
Dai, N. The diverse functions of IMP2/IGF2BP2 in metabolism. Trends Endocrinol. Metab. 31, 670–679 (2020).
Johann To Berens, P. & Molinier, J. Formation and recognition of UV-induced DNA damage within genome complexity. Int. J. Mol. Sci. 21, 6689 (2020).
Terman, B. I. et al. Identification of the KDR tyrosine kinase as a receptor for vascular endothelial cell growth factor. Biochem. Biophys. Res. Commun. 187, 1579–1586 (1992).
Yang, Y., **e, P., Opatowsky, Y. & Schlessinger, J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc. Natl Acad. Sci. USA 107, 1906–1911 (2010).
Leppänen, V.-M. et al. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc. Natl Acad. Sci. USA 107, 2425–2430 (2010).
Storz, J. F. High-altitude adaptation: mechanistic insights from integrated genomics and physiology. Mol. Biol. Evol. 38, 2677–2691 (2021).
Feng, Z., Cai, G. & Zheng, C. The mammals of **zang. 1–441 (Science Press, 1986).
Luo, Z. Rodentia Part III: Cricetidae. Fauna Sin. Mamm. 6, 121–128 (2000).
An, Z., John, E. K., Warren, L. P. & Stephen, C. P. Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 411, 62 (2001).
Guo, Z. et al. A major reorganization of Asian climate regime by the Early Miocene. Clim. Discuss. 4, 535–584 (2008).
Andersen, B. G. & Borns, H. W. The Ice Age World: An Introduction To Quaternary History And Research With Emphasis On North America And Northern Europe During The Last 2.5 Million Years. Vol. 33 (Scandinavian University Press, 1994).
Meng, L. et al. Refugial isolation and range expansions drive the genetic structure of Oxyria sinensis (Polygonaceae) in the Himalaya-Hengduan Mountains. Sci. Rep. 5, 10396 (2015).
McCormack, J. E., Huang, H., Knowles, L. L., Gillespie, R. & Clague, D. Sky islands. Encycl. Isl. 4, 841–843 (2009).
Deng, T. et al. Out of Tibet: pliocene woolly rhino suggests high-plateau origin of Ice Age megaherbivores. Science 333, 1285–1288 (2011).
Sikes, R. S. & Gannon, W. L. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J. Mammal. 92, 235–253 (2011).
Kaiser, H. F. & Rice, J. Little jiffy, mark IV. Educ. Psychol. Meas. 34, 111–117 (1974).
Bartlett, M. S. Properties of sufficiency and statistical tests. Proc. R. Soc. Lond. Ser. A-Math. Phys. Sci. 160, 268–282 (1937).
Abdi, H. & Williams, L. J. Tukey’s honestly significant difference (HSD) test. Encycl. Res. Des. 3, 1–5 (2010).
Conover, W. J. Practical Nonparametric Statistics. Vol. 350 (John Wiley & Sons, 1999).
Wei, T. et al. Package ‘corrplot’. Statistician 56, e24 (2017).
Wickham, H. ggplot2: Elegant Graphics For Data Analysis. (Springer, 2016).
Weisenfeld, N. I., Kumar, V., Shah, P., Church, D. M. & Jaffe, D. B. Direct determination of diploid genome sequences. Genome Res. 27, 757–767 (2017).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 12, 1–14 (2011).
Liu, S. et al. Ancient and modern genomes unravel the evolutionary history of the rhinoceros family. Cell 184, 4874–4885. e4816 (2021).
Meng, G., Li, Y., Yang, C. & Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47, e63–e63 (2019).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ar**v:1303.3997 (2013).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Esmukov, K. GeoPy, <https://github.com/geopy/geopy> (2022).
Kassambara, A. ggpubr:“ggplot2” based publication ready plots. R package version 0.1 7 (2018).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876 (2013).
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877 (2012).
Flouri, T., Jiao, X., Rannala, B. & Yang, Z. Species tree inference with BPP using genomic sequences and the multispecies coalescent. Mol. Biol. Evol. 35, 2585–2593 (2018).
Yang, Z. & Rannala, B. Bayesian species delimitation using multilocus sequence data. Proc. Natl Acad. Sci. USA 107, 9264–9269 (2010).
Rannala, B. & Yang, Z. Improved reversible jump algorithms for Bayesian species delimitation. Genetics 194, 245–253 (2013).
Yang, Z. & Rannala, B. Unguided species delimitation using DNA sequence data from multiple loci. Mol. Biol. Evol. 31, 3125–3135 (2014).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Zhang, C., Rabiee, M., Sayyari, E. & Mirarab, S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 19, 153 (2018).
Cole, T. L. et al. Genomic insights into the secondary aquatic transition of penguins. Nat. Commun. 13, 1–13 (2022).
Chifman, J. & Kubatko, L. Quartet inference from SNP data under the coalescent model. Bioinformatics 30, 3317–3324 (2014).
Swofford, D. L. & Sullivan, J. The Phylogenetic Handbook: A Practical Approach To Dna And Protein Phylogeny, Cáp. Vol. 7, 160–206 (Cambridge University Press, 2003).
Nguyen, L., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evolution 32, 268–274 (2015).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Kozlov, A. M., Aberer, A. J. & Stamatakis, A. ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics 31, 2577–2579 (2015).
Peters, S. E. & McClennen, M. The Paleobiology Database application programming interface. Paleobiology 42, 1–7 (2016).
Kumar, S., Stecher, G., Suleski, M. & Hedges, S. B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017).
Zhang, Y., Zheng, S. & Wei, G. Fossil arvicolines from the Leijiahe section, Lingtai, Gansu Province and current progress of Chinese arvicoline biochronology. Quat. Sci. 31, 622–635 (2011).
Lv, X. et al. Disjunct distribution and distinct intraspecific diversification of Eothenomys melanogaster in South China. BMC Evol. Biol. 18, 1–14 (2018).
Kohli, B. A., Fedorov, V. B., Waltari, E. & Cook, J. A. Phylogeography of a Holarctic rodent (Myodes rutilus): testing high latitude biogeographical hypotheses and the dynamics of range shifts. J. Biogeogr. 42, 377–389 (2015).
Bannikova, A. A. et al. Molecular phylogeny and evolution of the Asian lineage of vole genus Microtus (Rodentia: Arvicolinae) inferred from mitochondrial cytochrome b sequence. Biol. J. Linn. Soc. 99, 595–613 (2010).
Galewski, T. et al. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol. Biol. 6, 1–17 (2006).
Rambaut, A., Drummond, A. J., **e, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Field, A., Miles, J. & Field, Z. Discovering Statistics Using R (Sage publications, 2012).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Matzke, N. J. BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts. R. package, version 0. 2. 1, 2013 (2013).
Yang, C. et al. Evolutionary and biomedical insights from a marmoset diploid genome assembly. Nature 594, 227–233 (2021).
Bu, D. et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325 (2021).
Kiefer, F., Arnold, K., Künzli, M., Bordoli, L. & Schwede, T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 (2009).
Acknowledgements
We thank Rui Liao for assistance in collecting specimens in the field. Special thanks to Yinjuan Mao and Junhua Bai for drawing the figures. This work was supported by the National Natural Science Foundation of China (31470110, 31970399) and Survey of Wildlife Resources in Key Areas of Tibet (ZL202203601). This work was also supported by China National GeneBank (CNGB).
Author information
Authors and Affiliations
Contributions
Sy.L. and Sl.L. conceived the study. Sy.L. and Y.L. collected the samples. C.Z., G.M., T.W., M.T. and C.Y. performed bioinformatics and computational analyses of the data. R.W.M., Z.F., T.Z. and Y.Z. provided advice on the data analyses. Sy.L. and C.Z. drafted the manuscript. All authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
All authors have agreed to all manuscript contents, the author list and its order and the author contribution statements. All samples were obtained following the Guidelines of the American Society of Mammalogists and the laws and regulations of China for the implementation of the protection of terrestrial wild animals. Collecting protocols and research project were approved by the Ethics Committee of the Sichuan Academy of Forestry and the Institutional of Review Board on Bioethics and Biosafety of BGI (NO. FT17005).
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Luciano Matzkin and Luke R. Grinham.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, S., Zhou, C., Meng, G. et al. Evolution and diversification of Mountain voles (Rodentia: Cricetidae). Commun Biol 5, 1417 (2022). https://doi.org/10.1038/s42003-022-04371-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-04371-z
- Springer Nature Limited