Abstract
Comet assay is a widely used method, especially in the field of genotoxicity, to quantify and measure DNA damage visually at the level of individual cells with high sensitivity and efficiency. Generally, computer programs are used to analyze comet assay output images following two main steps. First, each comet region must be located and segmented, and next, it is scored using common metrics (e.g., tail length and tail moment). Currently, most studies on comet assay image analysis have adopted hand-crafted features rather than the recent and effective deep learning (DL) methods. In this paper, however, we propose a DL-based baseline method, called DeepComet, for comet segmentation. Furthermore, we created a trainable and testable comet assay image dataset that contains 1037 comet assay images with 8271 manually annotated comet objects. From the comet segmentation test results with the proposed dataset, the DeepComet achieves high average precision (AP), which is an essential metric in image segmentation and detection tasks. A comparative analysis was performed between the DeepComet and the state-of-the-arts automatic comet segmentation programs on the dataset. Besides, we found that the DeepComet records high correlations with a commercial comet analysis tool, which suggests that the DeepComet is suitable for practical application.
Similar content being viewed by others
Introduction
DNA damage is a major toxicological concern in drug development as it is a critical cause of cancer, as well as chronic and degenerative diseases such as heart diseases and Parkinson’s disease1,2,3,4,5,6. The DNA can be damaged by spontaneous chemical reactions, reactive oxygen, and nitrogen species generated by metabolism, or exogenous agents. If a damaged DNA is not repaired correctly, the damages might be accumulated and lead to mutations, cell death, or senescence that can induce various diseases including cancers and chronic diseases1. The assessment of DNA damage, therefore, is crucial in the toxicology field to detect genotoxicity and carcinogenic potential of certain compounds. It is also significant in medical research to understand the pathogenesis of diseases including cancer and chronic diseases, and can be applied in clinical medicine to monitor these disease conditions7,8.
Although there are several methods to assess DNA damage9,10,11,12, comet assay (also referred to as single-cell gel electrophoresis) is recommended for detecting the DNA strand breakage in ICH S2 (R1) guideline13. Its high sensitivity, high efficiency, and low cost made it easily accessible for researchers14,15. In the assay, damaged DNA migrates out of the nucleus to form a shape that resembles a tail of a comet during electrophoresis, whereas undamaged DNA in the nucleus forms the head of the comet shape. Followed by electrophoresis, DNA is visualized by staining with DNA-binding dye under fluorescence microscopy. If the comet head gets smaller and dimmer while its tail gets longer and brighter, it suggests that the DNA in the cell is significantly damaged. Hence, each cell detected in the image is referred to as a comet.
The damage of DNA on comet assay images can be determined either by visual scoring or computer image analysis16. Visual scoring classifies comets into several different categories based on DNA damage degree (visually estimated tail length and shape) subjectively. However, computer image analysis provides a range of different parameters of each comet by pixel-based computation. Although the visual scoring is simple and may appeal to those who cannot afford expensive computer programs, the computer image analysis is preferable in assessing DNA damage as it can provide objective quantified information of comets.
Several image analysis methods, such as Comet Score, CASP, OpenComet, CometQ, and HiComet for comet assay have been proposed to facilitate the comet scoring process16,17,18,19,20. They have two main stages, i.e., comet detection or segmentation and comet scoring. Depending on whether detection or segmentation process gets completed automatically, it can be divided into either a semi- or fully-automated tool. The primary performance differences come from whether a tool can detect and segment each comet correctly rather than scoring stage because the metric used to assess the comet score has predefined basic standards and rules21. However, these image analysis methods all use hand-crafted image features with traditional machine learning (such as support vector machine) at comet detection and segmentation stage. Users should define features manually and tune visually which might be laborious. It makes the methods have a few limitations on capturing comets from a noisy background and hardly distinguish two individual cells when overlapped. In contrast, DL can allow raw image data as input and learn to detect and segment comet in an end-to-end process.
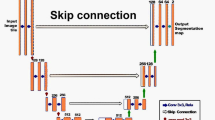
The DL provides excellent achievements in the fields of computer vision, such as image classification22, image object detection23,24, and image object segmentation5a). The terms involved are explained as follows:
Precision/recall curves of the DeepComet on the test dataset. (a) Intersection over Union (IoU) is given by the overlap** area between the predicted region and the ground truth region divided by the area of union of them. (b) The precision/recall curve of the DeepComet at IoU > 0.5 and IoU > 0.75 on our test dataset. The precision/recall curve can be used to evaluate the performance of an object detection model.
True positive (TP)
A detection where IoU ≥ threshold and the predicted class agrees with the ground truth class.
False positive (FP)
An incorrect detection.
False negative (FN)
A ground truth object not detected.
Precision
It measures relevant objects among all detected ones; it is expressed as follows.
Recall
It measures detected objects among all relevant ones; it is expressed as follows:
The AP is the precision averaged across all recall values between 0 and 1 by varying the detection confidence threshold. The COCO metric uses 101 interpolated data points to calculate the AP. In the following, APmask denotes that the IoU was estimated between segmentation masks, APbb indicates that the IoU was calculated between bounding boxes, mAP is the mean AP over all classes, AP@0.5 denotes that the IoU threshold was 0.5, and AP@[0.5:0.95] indicates that the AP was averaged over 10 IoU thresholds (from 0.5 to 0.95 with a step size of 0.05).
Figure 5b shows the precision/recall curve of the DeepComet at IoU > 0.5 and IoU > 0.75 on our test dataset. The precision/recall curve can be used to evaluate the performance of an object detection model as the confidence is changed by plotting a curve for each object class. A model is considered reliable if the curve stays close to y (precision) = 1, which means that the accuracy remains high as recall increases by varying the confidence threshold. For the segmentation of non-ghost and ghost cells, the DeepComet demonstrated that its precision remained over 0.9 until recall increased to approximately 0.8 at IoU > 0.5. The mAPmask@0.5 on the Normal test subset was 0.916, which is slightly higher than that of the Hard test dataset (0.87) and the total (Normal + Hard) test dataset (0.897). The results obtained on the Hard test dataset indicate that the DeepComet was well trained for segmenting comets on these noisy images. At IoU > 0.75, although the curves dropped as expected, the precision values were more significant than 0.8 until recall increased to approximately 0.8. The exception was when the recall value was over 0.4 with the Hard test dataset, where the precision values were slightly less than 0.8. Unlike at IoU > 0.5, there were gaps between the non-ghost curve and the ghost curve when IoU > 0.75.
In Table 2, we evaluated APmask and mAPmask averaged over IoU thresholds from 0.5 to 0.95 with a step size of 0.05. The mAPmask@[0.5:0.95] values of the Normal, Hard, and total datasets were 0.616, 0.584, and 0.601, respectively. In general, the DeepComet demonstrated a better performance in segmenting ghost cells compared to non-ghost cells on two datasets.
Table 3 compares the performance of the DeepComet with OpenComet Version.1.3.1 (https://www.cometbio.org)17 and HiComet (https://github.com/taehoonlee/HiComet)19. We applied default settings in each program. Since these two programs have no function to classify cells into non-ghost and ghost cells, we ignored the classification function of the DeepComet and simply transformed the output as detection of comets for comparison. The outputs of the HiComet are the detected bounding boxes of the comets. Thus, here the APbb metric was used to evaluate and compare the results. As shown in Table 3, the DeepComet showed much higher APbb values in all set-ups. The APbb@0.5 of our results were all greater than 0.9 on the three test datasets, whereas the results obtained using the OpenComet and HiComet were under 0.5. Regarding the high IoU threshold, the APbb@0.75 and APbb@[0.5:0.95] values of the DeepComet were approximately 0.78 and 0.66, respectively, whereas the compared programs barely reached 0.2. When comparing the corresponding APbb results obtained by the Normal and Hard test datasets, it was demonstrated that the gap between the results from each dataset was much less for the DeepComet compared to other methods, where DeepComet showed a difference of less than 5% but greater than 30% for others. However, it should be considered that the experimental setup can be unfavorable for OpenComet and HiComet as they were not optimized for our comet image dataset.
Figures 6 and 7 show the visual comparisons of the comet detection and segmentation results. Figure 6 shows the representative images from the Normal and Hard test datasets for comparison. The OpenComet could not segment slightly overlapped targets into individual comets. Furthermore, it did not effectively detect the entire tail of the comet for the segmentation No. 5 in the Normal image and No. 11 in the Hard image. There were also incorrect segmentations for ghost cells. From the HiComet output images in the third row, it was found that HiComet did not adequately recognize the tail part of the non-ghost cell and could not capture the entire ghost cell. However, the DeepComet in the bottom was able to segment slightly overlapped comets into two individuals. Also, the DeepComet captured a dim tail in the Normal image and a noisy tail in the Hard image. The DeepComet could also distinguish non-ghost and ghost cells correctly, and capture objects even on the edge of the image.
Figure 7 shows the segmentation results on representative comets, which prove challenging to segment due to overlap**, background noise, or a dim tail (bright background). Here, we added the results from Comet Assay IV Version 4.3 (Instem-Perceptive Instruments Ltd., Suffolk, Halstead, UK), which is a semi-automated method that requires a manual click on a comet to score. As shown in the first row, if two comets slightly overlap each other, the OpenComet and Comet Assay IV could not separate them and recognize each comet. In contrast, the HiComet only knew one of the comets. The second row shows the results on an image with background noise. Here, OpenComet never recognized the comet as valid, and Comet Assay IV detected the comet but could not segment it accurately, whereas the HiComet did it correctly. The images in the bottom have a bright background and a dim comet tail. The OpenComet and HiComet did not capture the entire tail, and the Comet Assay IV captured excessive area, including the background.
Comet score comparison
We validated our calculated comet scores by verifying the correlation with those from Comet Assay IV using randomly selected 107 non-ghost cells that could easily be segmented. The two most crucial comet parameters, DNA (%) in tail and Olive moment, were calculated, and the correlation between the scores obtained from the DeepComet and Comet Assay IV were analyzed using Pearson correlation. Figure 8 shows the correlations between the scores obtained for DeepComet and Comet Assay IV to be 0.917 for DNA (%) in tail and 0.959 for Olive moment. For both scores, the correlation was positive and significantly high.
In practical application, a fully-automated comet image analysis program should input sets of full comet assay images and automatically output all detected and segmented comets with their scores. Here, the comet scoring performance of the DeepComet was validated in comparison with Ground truth, Comet Assay IV, OpenComet, and HiComet. Figure 9 shows box plots of comet score (DNA (%) in tail and Olive moment) distributions of 50 comet assay images which were randomly selected from Normal and Hard test subsets (25 for each). The ground truth comet score distribution was calculated on manually annotated comet regions. The comet scores using Comet Assay IV were generated by manual click on every comet on the images using the program. For the objects that Comet Assay IV did not segment properly, we manually modified the comet regions as accurately as possible. False positive detection of comets which had no overlap** region with ground truth were all excluded. It is observed from the box plots that the results of our method match the ground truth the most, followed by Comet Assay IV which also shows quite similar shapes of box plots to the ground truth. OpenComet and HiComet detected almost all comets, however, the segmented comet tails were too short on our datasets, whereas Comet Assay IV tended to segment a comet tail a little wider and longer. Student’s t-test was carried out at the significance level of 0.05 to identify statistical difference between Ground truth and respective other methods. There was no significant difference between Ground truth and DeepComet as well as between Ground truth and Comet Assay IV, whereas there was a significant difference between Ground truth and OpenComet as well as between Ground truth and HiComet.
Discussion and conclusion
In this paper, we proposed DeepComet for comet segmentation that utilizes the Mask R-CNN architecture. Developed by Facebook AI research, the Mask R-CNN is one of the most popular architectures because it can be extended to other applications easily, such as human pose estimation. Its performance also has been verified in various practical applications44,45 and competitions (https://www.kaggle.com/competitions, https://github.com/matterport/Mask_RCNN) of image segmentation. Consequently, we selected the Mask R-CNN among different DL models for image object segmentationhttps://liveuou-my.sharepoint.com/:u:/g/personal/wooec52_liveuou_kr/EVIbAAmwXUBArB0B4uEmREMBe15FZTJB1LdU40KN8wwDUQ?e=i3jcnB).
There has been debate over the cause of ghost cells and how they should be analyzed appropriately. Due to their appearance, measurements of DNA (%) in the tail through image analysis are unreliable22. The DeepComet can classify cells into non-ghost and ghost cells, which helps users to apply customized measurements on ghost cells. With further post processing (e.g., background brightness subtraction, orientation correction, and head and tail separation), the comet score can be measured precisely for each segmented comet using the DeepComet.
In the precision/recall curve of DeepComet at IoU > 0.75, the precision values were higher than 0.8 until recall increased to approximately 0.8 except for the Hard test dataset, where the precision values were slightly lower than 0.8. Also, there were gaps between the curves of non-ghost and ghost cells. Based on visual observation, this occurred because the area of ghost cells was generally much more significant than non-ghost cells, which made IoU less sensitive to ghost cells. Furthermore, there were several non-ghost cells with very short and dim tails, which led to ambiguous annotations. The other experimental results for non-ghost and ghost cell segmentation demonstrated that the DeepComet obtained high mAPmask@0.5 (0.897) with the total test dataset. The results indicate that the DeepComet is superior to state-of-the-art products with comet bounding box detection using APbb. Besides, compared with the Normal test dataset, the APbb of the Hard test dataset gets reduced by approximately 30% for the state-of-the-arts, whereas the APbb of the DeepComet only decreased by less than 5%. In terms of comet scoring, our results proved that the DeepComet has a high correlation with the commercial Comet Assay IV program. Furthermore, the proposed method also shows the high performance when evaluated at a full comet assay image level. These points demonstrate that the DeepComet can be used for practical automatic comet assay image analysis.
Comet image variability of inter-laboratories is inevitable, and it could be a possible reason for the large gap in performance against our dataset between the DeepComet and other methods (as shown in Table 3 and Fig. 9). Comet images from different laboratories could vary in image resolution, background noise, comet shape, brightness, contrast, etc. As each method was developed and optimized using different datasets, DeepComet may also need to be validated on datasets other than ours. There should be datasets released from various places to carry out the more appropriate comparison, as well as to develop a more generalized algorithm.
We demonstrate that our newly proposed DL-based comet assay analysis program, DeepComet, has shown great performance on our publicly released datasets. The DeepComet and our datasets can serve as a baseline for comet image segmentation and analysis to facilitate future research in this field of toxicology and medical science. In the future, we hope to expand the datasets with multiple external sources from different experimental environments and develop a more specific and accurate deep learning-based architecture for comet assay image analysis.
Data availability
The datasets generated during and/or analyzed during the current study are available here: https://doi.org/10.5281/zenodo.6395303.
Change history
19 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-10754-3
References
Hoeijmakers, J. H. DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 (2009).
Kadioglu, E., Sardas, S., Aslan, S., Isik, E. & Karakaya, A. E. Detection of oxidative DNA damage in lymphocytes of patients with Alzheimer’s disease. Biomarkers 9, 203–209 (2004).
Psimadas, D. et al. DNA damage and repair efficiency in lymphocytes from schizophrenic patients. Cancer Lett. 204, 33–40 (2004).
Collins, A. R. et al. DNA damage in diabetes: Correlation with a clinical marker. Free Radical Biol. Med. 25, 373–377 (1998).
Andreazza, A. C. et al. DNA damage in bipolar disorder. Psychiatry Res. 153, 27–32 (2007).
Kopjar, N., Garaj-Vrhovac, V. & Milas, I. Assessment of chemotherapy-induced DNA damage in peripheral blood leukocytes of cancer patients using the alkaline comet assay. Teratog. Carcinog. Mutagen. 22, 13–30 (2002).
Gunasekarana, V., Raj, G. V. & Chand, P. A comprehensive review on clinical applications of comet assay. J. Clin. Diagn. Res. JCDR 9, GE01–GE05 (2015).
Møller, P., Stopper, H. & Collins, A. R. Measurement of DNA damage with the comet assay in high-prevalence diseases: Current status and future directions. Mutagenesis 35, 5–18 (2020).
Quinet, A., Carvajal-Maldonado, D., Lemacon, D. & Vindigni, A. DNA fiber analysis: Mind the gap!. Methods Enzymol. 591, 55–82 (2017).
Gassman, N. R. & Wilson, S. H. Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair 31, 52–63 (2015).
Furda, A., Santos, J. H., Meyer, J. N. & Van Houten, B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 1105, 419–437 (2014).
Baumstark-Khan, C., Hentschel, U., Nikandrova, Y., Krug, J. & Horneck, G. Fluorometric analysis of DNA unwinding (FADU) as a method for detecting repair-induced DNA strand breaks in UV-irradiated mammalian cells. Photochem. Photobiol. 72, 477–484 (2000).
ICH Guideline. Guidance on genotoxicity testing and data interpretation for pharmaceuticals intended for human use S2 (R1). In International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, Rockville, Maryland, US (2011).
Singh, N. P., McCoy, M. T., Tice, R. R. & Schneider, E. L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175, 184–191 (1988).
Ostling, O. & Johanson, K. J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123, 291–298 (1984).
Ganapathy, S., Muraleedharan, A., Sathidevi, P. S., Chand, P. & Rajkumar, R. P. CometQ: An automated tool for the detection and quantification of DNA damage using comet assay image analysis. Comput. Methods Programs Biomed. 133, 143–154 (2016).
Gyori, B. M., Venkatachalam, G., Thiagarajan, P., Hsu, D. & Clement, M.-V. OpenComet: An automated tool for comet assay image analysis. Redox Biol. 2, 457–465 (2014).
Końca, K. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 534, 15–20 (2003).
Lee, T. et al. HiComet: A high-throughput comet analysis tool for large-scale DNA damage assessment. BMC Bioinform. 19, 44 (2018).
Helma, C. & Uhl, M. A public domain image-analysis program for the single-cell gel-electrophoresis (comet) assay. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 466, 9–15 (2000).
Kumaravel, T., Vilhar, B., Faux, S. P. & Jha, A. N. Comet assay measurements: A perspective. Cell Biol. Toxicol. 25, 53–64 (2009).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 1097–1105 (2012).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: Unified, real-time object detection. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 779–788 (2016).
Ren, S., He, K., Girshick, R. & Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Advances in Neural Information Processing Systems 91–99 (2015).
Chen, L.-C., Papandreou, G., Schroff, F. & Adam, H. Rethinking atrous convolution for semantic image segmentation. http://ar**v.org/ar**v:1706.05587 (2017).
Zhao, H., Shi, J., Qi, X., Wang, X. & Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2881–2890 (2017).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. In 2017 IEEE International Conference on Computer Vision (ICCV) 2980–2988 (2017).
Russakovsky, O. et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015).
LeCun, Y., Cortes, C. & Burges, C. J. The MNIST database of handwritten digits https://yann.lecun.com/exdb/mnist (2009).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition. 248–255 (2009).
Lin, T.-Y. et al. Microsoft COCO: Common Objects in Context. In Computer Vision—ECCV 2014.(eds Fleet, D., Pajdla, T., Schiele, B., & Tuytelaars, T.) 740–755 (Springer, 2014).
Turan, M. K. & Sehirli, E. A novel method to identify and grade DNA damage on comet images. Comput. Methods Programs Biomed. 147, 19–27 (2017).
Atila, Ü., Baydilli, Y. Y., Sehirli, E. & Turan, M. K. Classification of DNA damages on segmented comet assay images using convolutional neural network. Comput. Methods Programs Biomed. 186, 105192 (2020).
Afiahayati, E. A., Yanuaryska, R. D., Nuha, F. U. & Mulyana, S. Comet assay classification for Buccal Mucosa’s DNA damage measurement with super tiny dataset using transfer. Intell. Inf. Database Syst. Rec. Dev. 830, 279 (2019).
Dutta, A. & Zisserman, A. The VIA annotation software for images, audio and video. In Proceedings of the 27th ACM International Conference on Multimedia. 2276–2279 (2019).
Lorenzo, Y., Costa, S., Collins, A. R. & Azqueta, A. The comet assay, DNA damage, DNA repair and cytotoxicity: Hedgehogs are not always dead. Mutagenesis 28, 427–432 (2013).
OECD. Test No 489: In vivo Mammalian alkaline Comet assay. In OECD Guidelines for the Testing of Chemicals. OECD Publ Vol. 4, 1–21 (2014).
Lin, T. et al. Feature pyramid networks for object detection. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 936–944 (2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (2016).
Hellman, B., Vaghef, H. & Boström, B. The concepts of tail moment and tail inertia in the single cell gel electrophoresis assay. Mutat. Res. DNA Repair 336, 123–131 (1995).
Kumaravel, T. & Jha, A. N. Reliable Comet assay measurements for detecting DNA damage induced by ionising radiation and chemicals. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 605, 7–16 (2006).
Olive, P. L., Banáth, J. P. & Durand, R. E. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the" comet" assay. Radiat. Res. 122, 86–94 (1990).
Everingham, M., Van Gool, L., Williams, C. K., Winn, J. & Zisserman, A. The Pascal visual object classes (VOC) challenge. Int. J. Comput. Vis. 88, 303–338 (2010).
Anantharaman, R., Velazquez, M. & Lee, Y. Utilizing Mask R-CNN for Detection and Segmentation of Oral Diseases. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2197–2204 (2018).
Pobar, M. & Ivasic-Kos, M. Mask R-CNN and Optical Flow Based Method for Detection and Marking of Handball Actions. In 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) 1–6 (2018).
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No. 2019M3E5D4066897).
Author information
Authors and Affiliations
Contributions
W.-C.S. and I.S. conceived and designed the study. Y. H., H.-J.H., H.L., and D.L performed material preparation, data collection, and analysis. J.K., Z.-y.H., J.-Y.L., J.-H.S., and H.S.L. helped data collection with annotation on images. The first and second drafts, and the revised version of the manuscript were written by Y.H. and H.-J.H. All authors commented on the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original online version of this Article included an incorrect link to access the data in the Data Availability section where “The datasets generated during and/or analyzed during the current study are available in the following domain, https://liveuoumy.sharepoint.com/:u:/g/personal/wooec52_liveuou_kr/EVIbAAmwXUBArB0B4uEmREMBe15FZTJB1LdU40KN8wwDUQ?e=i3jcnB”. Now reads “The datasets generated during and/or analyzed during the current study are available here: https://doi.org/10.5281/zenodo.6395303”.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hong, Y., Han, HJ., Lee, H. et al. Deep learning method for comet segmentation and comet assay image analysis. Sci Rep 10, 18915 (2020). https://doi.org/10.1038/s41598-020-75592-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75592-7
- Springer Nature Limited
This article is cited by
-
Exploring the Impact of Simulated Microgravity on Cellular DNA: A Comparative Analysis of Cancer and Normal Cell Lines
Microgravity Science and Technology (2024)
-
Arsenic Induced Oxidative Neural-Damages in Rat are Mitigated by Tea-Leave Extract via MMPs and AChE Inactivation, Shown by Molecular Docking and in Vitro Studies with Pure Theaflavin and AChE
Cell Biochemistry and Biophysics (2024)