Abstract
Chinese sturgeon (Acipenser sinensis) has been listed as a critically endangered species on the IUCN Red List and is an endemic fish of China. Five sets of duplex polymerase chain reactions (PCR) assays were developed with 10 tetranucleotide microsatellites for Chinese sturgeon. The size of CS57, ZHX43, ZHX69, AS105, ZHX51, AS074, ZHX2, AS078, AS026 and AS073 products in 184 Chinese sturgeon individuals ranged from 257–305, 191–241, 251–285, 172–244, 236–260, 169–209, 194–234, 92–176, 165–257 and 120–164, respectively. The observed allele number of the 10 microsatellites ranged from 7 to 16, and the total number of alleles was 106. The number of alleles per individual in CS57, ZHX43, AS105, AS074, AS078 and AS026 was 1–4. The number of alleles per individual in ZHX69, ZHX51, ZHX2 and AS073 was 2–4. The mean number of alleles per locus per individual ranged from 2.01–3.76. The expected heterozygosity (HE), observed heterozygosity (HO), polymorphic information content (PIC) and Shannon-Weiner diversity index (H′) ranged from 0.582 to 0.899, from 0.676 to 1, from 0.518 to 0.886 and from 1.034 to 2.34, respectively. Despite many advantages, the use of microsatellites as genetic analysis tools can be limited by the cost of the associated experiment. To solve this problem, this set of five duplex PCRs will provide tools that are more helpful, less expensive and less time consuming than others used for genetic analyses in Chinese sturgeon.
Similar content being viewed by others
Introduction
Chinese sturgeon (Acipenser sinensis) is an endemic and critically endangered species in China. Chinese sturgeon was once widely distributed in the Yangtze River, Pearl River and Chinese seas1. The wild population of Chinese sturgeon has fallen drastically in the past decades due to human activities, such as the use of pesticides, industrial overfishing and environmental pollution2,3,4. Currently, the wild population of Chinese sturgeon in the Yangtze River is very small. The need for species rehabilitation is known, and efforts have been made to protect it. To prevent this species from becoming extinct, artificial propagation and tagged ranching have been performed every year since 19845,24. However, by using trinucleotide or tetranucleotide microsatellites instead of dinucleotide microsatellites, the risk of obtaining false alleles is reduced25. Studies have shown that four-base microsatellites are more stable and more accurate than other types of microsatellites26. Furthermore, PCR amplification results from tetranucleotide repeat loci are easier to interpret than in the case of dinucleotide repeat loci because only a single stutter band is typically observed, in a position four bases shorter than each allele band, and the intensity of the stutter band is generally <10% of the main band27. Currently, the human and bovine paternity test kits used worldwide consist of four-base microsatellite markers. Multiplex PCR refers to the simultaneous amplification of multiple microsatellite loci in a PCR system, resulting in multiple PCR amplification products. Multiplex PCR technology has been widely employed in many aquatic animals28,29. This technology is not only greatly improving the efficiency of genoty** but also reducing the cost of genetic analyses. Compared with the conventional simplex procedure, multiplex SSR sets and multi-loading combinations reduced the costs of the PCR related reagents by about 50% and the costs of the electrophoresis related reagents by over 85%30.
In this study we provide five duplex PCR assays performed with 10 tetranucleotide microsatellites in order to help in the traceability of individuals and in the genetic conservation management of Chinese sturgeon.
Results
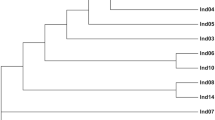
A total of 412 sequences that contain tetranucleotide microsatellites were selected from the cDNA libraries. And 412 novel primer pairs were developed using Primer Premier 5.0 software. Of the 412 microsatellites that were tested with the DNA of 12 individuals, 315 resulted in no or poor amplification in Chinese sturgeon. Eighty-seven microsatellites produced ambiguous polymorphic products. The remaining 10 microsatellites were successfully amplified. Among the 31 tetranucleotide microsatellites reported by Zhu B14. that were tested with the DNA of 12 individuals, only 12 microsatellites showed polymorphism. The rest of the microsatellites showed ambiguous polymorphic products. According to the results for these microsatellites tested in the 184 individuals, we developed five novel polymorphic loci (CS57, ZHX43, ZHX69, ZHX51 and ZHX2) and selected five polymorphic loci (AS105, AS074, AS078, AS026 and AS073) previously reported in Chinese sturgeon to build five duplex PCR assays (Table 1). Sequences containing these five microsatellite markers that we developed were deposited in GenBank under accession numbers MN401754-MN401758. The MPprimer software showed no pairs of loci displayed significant linkage disequilibrium. The size of the products for the ten microsatellites in 184 Chinese sturgeon individuals has been listed in the Table 2. The observed allele number of the 10 microsatellites ranged from 7 to 16, and the total number of alleles was 106. The number of alleles per individual in CS57, ZHX43, AS105, AS074, AS078 and AS026 was 1–4. The number of alleles per individual in ZHX69, ZHX51, ZHX2 and AS073 was 2–4. The mean number of alleles per locus per individual ranged from 2.01–3.76. The expected heterozygosity (HE), observed heterozygosity (HO), polymorphic information content (PIC) and Shannon-Weiner diversity indices (H′) ranged from 0.582 to 0.899, from 0.676 to 1, from 0.518 to 0.886 and from 1.034 to 2.34, respectively (Table 2). The frequencies of null alleles ranging from 0.03 to 0.15 were reported by the program MICROCHECKER (Table 2). The 184 Chinese sturgeon individuals were accurately reconstructed in the UPGMA dendrogram using the ten microsatellites (Fig. 1).
Discussion
Microsatellites are very important for the management and conservation of fish species31. Although many microsatellites have been developed in Chinese sturgeonEstablishing the microsatellite based duplex PCR assays for population studies of Chinese sturgeon According to the size of PCR products in 184 individuals, we obtain the range of sizes for each selected microsatellite. Duplex PCR site combinations were chosen according to the range of sizes for each selected microsatellite and the avoidance of potential hairpin structures and primer dimers. Every duplex PCR assay reaction in a total volume of 25 μl contained 0.25 U of Taq polymerase (Takara, China), 0.25 μM each primer, about 50–100 ng template DNA, 0.25 μM dNTPs, 1.5 mM MgCl2, 0.25 μM PCR buffer (Takara, China) and water. PCR was performed under the following profile: an initial step at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, and an extension at 72 °C for 10 min. The PCR products were size fractionated with 10% PAGE and visualized by silver staining. The sizes of the alleles were estimated with the pBR322 DNA/Mspl marker (Takara). The statistics of the polymorphic parameters, including the mean expected heterozygosity (HE), observed heterozygosity (HO) and Shannon-Weiner diversity indices (H′) were calculated using ATetra1.249 software. The polymorphic information content (PIC) was calculated using the formula \(PIC=1-\mathop{\sum }\limits_{i=1}^{n}{P}_{i}^{2}-\mathop{\sum }\limits_{i=1}^{n-1}\mathop{\sum }\limits_{j=i+1}^{n}2{P}_{i}^{2}{P}_{j}^{2}\), Pi and Pj are the frequencies of I and J allele in the microsatellite loci. The presence of null alleles was assessed at a 95% confidence interval using the program MICROCHECKER50. The software MPprimers51 was used to detect whether the duplex PCR primers in this study were in linkage disequilibrium. The ten microsatellite loci were scored in a presence/absence format, so that genotypic data were transformed into allele phenotypes52. Each phenotype classified alleles as present or absent, regardless of dose. Based on the allele phenotypes, a dendrogram was constructed using the unweighted pair group arithmetic means (UPGMA) clustering algorithm in PHYLIP’S NEIGHBOR version 3.6953. The constructed tree file was visualized using MEGA version 5.154.Genetic analysis
Data availability
Data are available from the corresponding author upon reasonable request.
References
Yang, D. et al. Distribution and movement of Chinese sturgeon, Acipenser sinensis, on the spawning ground located below the Gezhouba Dam during spawning seasons. J. Appl. Ichthyol. 22, 145–151 (2010).
Wu, J. M. et al. Drastic decline in spawning activity of Chinese sturgeon Acipenser sinensis Gray 1835 in the remaining spawning ground of the Yangtze River since the construction of hydrodams. J. Appl. Ichthyol. 31, 839–842 (2015).
Besser, J. M., Wang, N., Dwyer, F. J., Mayer, F. L. Jr. & Ingersoll, C. G. Assessing contaminant sensitivity of endangered and threatened aquatic species: part II. Chronic toxicity of copper and pentachlorophenol to two endangered species and two surrogate species. Arch. Environ. Contam. Toxicol. 48, 155–165 (2005).
Wang, J. H., Wei, Q. W. & Zou, Y. C. Conservation strategies for the Chinese sturgeon, Acipenser sinensis: an overview on 30 years of practices and future needs. J. Appl. Ichthyol. 27, 176–180 (2011).
Fu, Z. L., Lu, D. C. & Chen, S. L. The artificial propagated Chinese sturgeon below the Gezhouba Dam. Freshw. Fish. 1, 1–5 (1985).
**ao, H., Chang, J. & Liu, Y. Evaluation on status of artificial propagation and releasing of Chinese sturgeon in the Yangtze River. Acta Hydrobiol. Sin. 23, 572–576 (1999).
Wei, Q. W. Acipenser sinensis. The IUCN Red List of Threatened Species 2010: e.T236A13044272, https://doi.org/10.2305/IUCN.UK.2010-1.RLTS.T236A13044272.en (2010).
Yue, P. Q. & Chen, Y. Y. China Red Data Book of Endangered Animals: Pisces (ed. Wang, S., Le, P. Y. & Chen, Y. Y.) 13–16 (Science Press, 1998).
Kohlmann, K. et al. Validation of 12 species-specific, tetrasomic microsatellite loci from the Russian sturgeon, Acipenser gueldenstaedtii, for genetic broodstock management. Aquacult. Int. 26, 1365 (2018).
Kohlmann, K. et al. Isolation and characterization of 18 polymorphic microsatellite markers for the critically endangered stellate sturgeon, Acipenser stellatus. Eur. J. Wildl. Res. 63, 75 (2017).
Boscari, E. et al. Microsatellites from the genome and the transcriptome of the tetraploid Adriatic sturgeon, Acipenser naccarii (Bonaparte, 1836) and cross-species applicability to the diploid beluga sturgeon, Huso huso (Linnaeus, 1758). J. Appl. Ichthyol. 31, 977–983 (2015).
Que, Y. et al. Identification and characterization of seventeen novel microsatellite markers for Dabry’s sturgeon (Acipenser dabryanus). J. Genet. 94, 62 (2015).
**n, M. M. et al. Twenty-four novel microsatellites for the endangered Chinese sturgeon (Acipenser sinensis Gray, 1835). J. Appl. Ichthyol. 32, 405–408 (2016).
Zhu, B., Liao, X., Shao, Z., Rosenthal, H. & Chang, J. Isolation and characterization of microsatellites in Chinese sturgeon, Acipenser sinensis. Mol. Ecol. Notes. 5, 888–892 (2005).
Zhu, B. et al. Analysis of genetic variation in the Chinese sturgeon, Acipenser sinensis: estimating the contribution of artificially produced larvae in a wild population. J. Appl. Ichthyol. 18, 301–306 (2002).
Wang, J. H., Wei, Q. W. & Zou, Y. C. Conservation strategies for the Chinese sturgeon, Acipenser sinensis: an overview on 30° years of practices and future needs. J. Appl. Ichthyol. 27, 176–180 (2011).
Tautz, D. & Renz, M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 12, 4127–4138 (1984).
O’Connell, M. & Wright, J. M. Microsatellite DNA in fishes. Rev. Fish. Biol. Fish. 7, 331–363 (1997).
Congiu, L. et al. Managing polyploidy in ex situ conservation genetics: the case of the critically endangered Adriatic sturgeon (Acipenser naccarii). PLoS One. 6, e18249 (2011).
Luo, W. et al. Rapid Development of Microsatellite Markers for the Endangered Fish Schizothorax biddulphi (Günther) Using Next Generation Sequencing and Cross-Species Amplification. Int. J. Mol. Sci. 13, 14946–14955 (2012).
Liu, X., Luo, W., Zeng, C., Wang, W. & Gao, Z. Isolation of New 40 Microsatellite Markers in Mandarin Fish (Siniperca chuatsi). Int. J. Mol. Sci. 12, 4180–4189 (2011).
Wang, H., Luo, X., Shi, W. B. & Zhang, B. W. Development and characterization of fourteen novel microsatellite loci in Chinese muntjac (Muntiacus reevesi). Conserv. Genet. Resour. 5, 1083–1085 (2013).
Archie, E. A., Moss, C. J. & Alberts, S. C. Characterization of tetranucleotide microsatellite loci in the African Savannah Elephant (Loxodonta africana africana). Mol. Ecol. Notes. 3, 244–246 (2003).
Taberlet, P. et al. Reliable genoty** of samples with very low DNA quantities using PCR. Nucleic. Acids. Res. 24, 3189–3194 (1996).
Taberlet, P., Waits, L. P. & Luikart, G. Noninvasive genetic sampling: Look before you leap. Trends. Ecol. Evol. 14, 323–327 (1999).
Walsh, P. S., Fildes, N. J. & Reynolds, R. Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic. Acids. Res. 24, 2807–2812 (1996).
Urquhart, A., Oldroyd, N. J., Kimpton, C. P. & Gill, P. Highly discriminating heptaplex short tandem repeat PCR system for forensic identification. BioTechniques 18, 116–121 (1995).
Yan, W., Wang, X., Wang, A. & Guo, X. A 16-microsatellite multiplex assay for parentage assignment in the eastern oyster (Crassostrea virginica Gmelin). Aquaculture. 308, S28–S33 (2010).
Li, Y. et al. Development of two microsatellite multiplex systems for black tiger shrimp Penaeus monodon and its application in genetic diversity study for two populations. Aquaculture. 266, 279–288 (2007).
Merdinoglu, D. et al. Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol. Breed. 15, 349–366 (2005).
Moghim, M. et al. Application of microsatellite markers for genetic conservation and management of Persian sturgeon (Acipenser persicus, Borodin, 1897) in the Caspian Sea. J. Appl. Ichthyol. 29, 696–703 (2013).
Rajkov, J., Shao, Z. J. & Berrebi, P. Evolution of polyploidy and functional diploidization in sturgeons: Microsatellite analysis in 10 sturgeon species. J. Hered. 105, 521–531 (2014).
González-Castellano, I., Perina, A., González-Tizón, A. M., Torrecilla, Z. & Martínez-Lage, A. Isolation and characterization of 21 polymorphic microsatellite loci for the rockpool shrimp Palaemon elegans using Illumina MiSeq sequencing. Sci. Rep. 8, 17197 (2016).
Meirmans, P. G., Liu, S. L. & Tienderen, P. H. The Analysis of Polyploid Genetic Data. J. Hered. 109, 283–296 (2018).
Waples, R. S. Testing for Hardy–Weinberg proportions: have we lost the plot? J. Hered. 106, 1–19 (2015).
Nei, M. Molecular evolutionary genetics. New York: Columbia University Press (1987).
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292 (1972).
Hedrick, P. W. A standardized genetic differentiation measure. Evolution. 59, 1633–1638 (2005).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics. 155, 945–959 (2000).
Wang, J. & Scribner, K. T. Parentage and sibship inference from markers in polyploids. Mol. Ecol. Resour. 14, 541–553 (2014).
Li, P. et al. Development of eighteen tetranucleotide microsatellite markers in Tibetan macaque (Macaca thibetana) and genetic diversity analysis of captive population. Biochem. Syst. Ecol. 57, 293–296 (2014).
Chambers, G. K. & MacAvoy, E. S. Microsatellites: consensus and controversy. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126, 455–476 (2000).
Nater, A., Kopps, A. M. & Krützen, M. New polymorphic tetranucleotide microsatellite improve scoring accuracy in the bottlenose dolphin Tursiops aduncus. Mol. Ecol. Resour. 9, 531–534 (2009).
Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7, 1–6 (2018).
Du, H. J. et al. Hypothalamus-pituitary-gonad axis transcriptome profiling for sex differentiation in Acipenser sinensis. Sci. Data 6, 87 (2019).
Pertea, G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 19, 651–652 (2003).
Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33, 2583 (2017).
Lalitha, S. Primer Premier 5. Biotech. Softw. Internet Rep. 1, 270–272 (2000).
Puyvelde, K. V., Geert, A. V. & Triest, L. Atetra, a new software program to analyse tetraploid microsatellite data: comparison with tetra and tetrasat. Mol. Ecol. Resour. 10, 331–334 (2010).
Van, O. C., Hutchinson, W. F., Wills, D. P. M. & Shipley, P. Micro-checker: software for identifying and correcting genoty** errors in microsatellite data. Mol. Ecol. Notes. 4, 535–538 (2004).
Shen, Z. et al. MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics. 11, 143 (2010).
Becher, S. A. et al. Microsatellites for cultivar identification in Pelargonium. THEOR. APPL. Genet. 101, 643–651 (2000).
Felsenstein, J. Phylip, Phylogeny Inference Package (version 3.6.6). Retrived from, http://evolution.genetics.washington.edu/phylip.html (2005)
Kumar, S., Tamura, K. & Nei, M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004).
Acknowledgements
This work was supported by the Three Gorges Environment Protection Fund, Chinese Three Gorges Corporation (no, XN270).
Author information
Authors and Affiliations
Contributions
Y.H., H.D. and X.L. conceived and designed the research. Y.H. wrote the manuscript with contributions from H.D. and J.Y. B.W. analysed the data. Y.H., K.X. and X.L. carried out the experiment.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hu, Y., Liu, X., Yang, J. et al. Development and characterization of a duplex PCR assay in Chinese sturgeon (Acipenser sinensis) for genetic analysis. Sci Rep 10, 3451 (2020). https://doi.org/10.1038/s41598-020-60401-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60401-y
- Springer Nature Limited