Abstract
The EMDataResource Ligand Model Challenge aimed to assess the reliability and reproducibility of modeling ligands bound to protein and protein–nucleic acid complexes in cryogenic electron microscopy (cryo-EM) maps determined at near-atomic (1.9–2.5 Å) resolution. Three published maps were selected as targets: Escherichia coli beta-galactosidase with inhibitor, SARS-CoV-2 virus RNA-dependent RNA polymerase with covalently bound nucleotide analog and SARS-CoV-2 virus ion channel ORF3a with bound lipid. Sixty-one models were submitted from 17 independent research groups, each with supporting workflow details. The quality of submitted ligand models and surrounding atoms were analyzed by visual inspection and quantification of local map quality, model-to-map fit, geometry, energetics and contact scores. A composite rather than a single score was needed to assess macromolecule+ligand model quality. These observations lead us to recommend best practices for assessing cryo-EM structures of liganded macromolecules reported at near-atomic resolution.




Similar content being viewed by others
Data availability
Cryo-EM map targets were the primary maps of EMDB entries EMD-7770, EMD-30210 and EMD-22898 (www.ebi.ac.uk/emdb, emdataresource.org). Reference models were PDB entries 6CVM v.1.3 (target 1), 7BV2 v.3.4 (target 2) and 7KJR v.1.1 (target 3) wwpdb.org. Submitted models, model metadata, result logs and compiled data are available via challenges.emdataresource.org/?q=2021-model-challenge and archived via Zenodo at https://doi.org/10.5281/zenodo.10551958 (ref. 90). Interactive summary tables, graphical views and spreadsheet downloads of compiled results are available at model-compare.emdataresource.org/2021/cgi-bin. Source Data are provided with this paper.
References
Adams, P. D. et al. Outcome of the First wwPDB/CCDC/D3R Ligand Validation Workshop. Structure 24, 502–508 (2016).
Gore, S. et al. Validation of structures in the Protein Data Bank. Structure 25, 1916–1927 (2017).
Smart, O. S. et al. Validation of ligands in macromolecular structures determined by X-ray crystallography. Acta Crystallogr. D. Struct. Biol. 74, 228–236 (2018).
Feng, Z. et al. Enhanced validation of small-molecule ligands and carbohydrates in the Protein Data Bank. Structure 29, 393–400.e1 (2021).
Lawson, C. L., Berman, H. M. & Chiu, W. Evolving data standards for cryo-EM structures. Struct. Dyn. 7, 014701 (2020).
Lawson, C. L. & Chiu, W. Comparing cryo-EM structures. J. Struct. Biol. 204, 523–526 (2018).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Lawson, C. L. et al. Cryo-EM model validation recommendations based on outcomes of the 2019 EMDataResource challenge. Nat. Methods 18, 156–164 (2021).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020).
Wang, Z., Patwardhan, A. & Kleywegt, G. J. Validation analysis of EMDB entries. Acta Crystallogr. D. Struct. Biol. 78, 542–552 (2022).
Bartesaghi, A. et al. Atomic resolution cryo-EM structure of β-Galactosidase. Structure 26, 848–856.e3 (2018).
Yin, W. et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 (2020).
Kokic, G. et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 12, 279 (2021).
Kern, D. M. et al. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 28, 573–582 (2021).
Kryshtafovych, A., Adams, P. D., Lawson, C. L. & Chiu, W. Evaluation system and web infrastructure for the second cryo-EM model challenge. J. Struct. Biol. 204, 96–108 (2018).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Lagerstedt, I. et al. Web-based visualisation and analysis of 3D electron-microscopy data from EMDB and PDB. J. Struct. Biol. 184, 173–181 (2013).
Joseph, A. P., Lagerstedt, I., Patwardhan, A., Topf, M. & Winn, M. Improved metrics for comparing structures of macromolecular assemblies determined by 3D electron-microscopy. J. Struct. Biol. 199, 12–26 (2017).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D. Struct. Biol. 74, 814–840 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Kryshtafovych, A. et al. Challenging the state of the art in protein structure prediction: highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10. Proteins 82, 26–42 (2014).
Bruno, I. J. et al. Retrieval of crystallographically-derived molecular geometry information. J. Chem. Inf. Comput. Sci. 44, 2133–2144 (2004).
Shao, C. et al. Simplified quality assessment for small-molecule ligands in the Protein Data Bank. Structure 30, 252–262.e4 (2022).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1069–1078 (2020).
Nicholls, R. A. et al. Modelling covalent linkages in CCP4. Acta Crystallogr. D. Struct. Biol. 77, 712–726 (2021).
Černý, J., Božíková, P., Svoboda, J. & Schneider, B. A unified dinucleotide alphabet describing both RNA and DNA structures. Nucleic Acids Res. 48, 6367–6381 (2020).
Černý, J. et al. Structural alphabets for conformational analysis of nucleic acids available at dnatco.datmos.org. Acta Crystallogr. D. Struct. Biol. 76, 805–813 (2020).
Biedermannová, L. & Schneider, B. Structure of the ordered hydration of amino acids in proteins: analysis of crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 71, 2192–2202 (2015).
Černý, J., Schneider, B. & Biedermannová, L. WatAA: Atlas of Protein Hydration. Exploring synergies between data mining and ab initio calculations. Phys. Chem. Chem. Phys. 19, 17094–17102 (2017).
Prisant, M. G., Williams, C. J., Chen, V. B., Richardson, J. S. & Richardson, D. C. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink ‘waters,’ and NGL Viewer to recapture online 3D graphics. Protein Sci. 29, 315–329 (2020).
Jiang, S., Feher, M., Williams, C., Cole, B. & Shaw, D. E. AutoPH4: an automated method for generating pharmacophore models from protein binding pockets. J. Chem. Inf. Model. 60, 4326–4338 (2020).
Tyagi, R., Singh, A., Chaudhary, K. K. & Yadav, M. K. in Bioinformatics (eds Singh, D. B. & Pathak, R. K.) Ch. 17, 269–289 (Academic, 2022).
Sellers, B. D., James, N. C. & Gobbi, A. A comparison of quantum and molecular mechanical methods to estimate strain energy in druglike fragments. J. Chem. Inf. Model. 57, 1265–1275 (2017).
Lee, M.-L. et al. chemalot and chemalot_knime: command line programs as workflow tools for drug discovery. J. Cheminform. 9, 38 (2017).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Croll, T. I., Williams, C. J., Chen, V. B., Richardson, D. C. & Richardson, J. S. Improving SARS-CoV-2 structures: peer review by early coordinate release. Biophys. J. 120, 1085–1096 (2021).
Modi, V., Xu, Q., Adhikari, S. & Dunbrack, R. L. Jr. Assessment of template-based modeling of protein structure in CASP11. Proteins 84, 200–220 (2016).
Giri, N. & Cheng, J. Improving protein-ligand interaction modeling with cryo-EM data, templates, and deep learning in 2021 Ligand Model Challenge. Biomolecules 13, 132 (2023).
Zhang, K. et al. Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. 10, 5511 (2019).
Su, Z. et al. Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 Å resolution. Nature 596, 603–607 (2021).
Lawson, C. L., Berman, H. M., Chen, L., Vallat, B. & Zirbel, C. L. The Nucleic Acid Knowledgebase: a new portal for 3D structural information about nucleic acids. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad957 (2023).
Sun, S. Y. et al. Cryo-ET of parasites gives subnanometer insight into tubulin-based structures. Proc. Natl Acad. Sci. USA 119, e2111661119 (2022).
Liu, H.-F. et al. nextPYP: a comprehensive and scalable platform for characterizing protein variability in situ using single-particle cryo-electron tomography. Nat. Methods 20, 1909–1919 (2023).
Chmielewski, D. et al. Structural insights into the modulation of coronavirus spike tilting and infectivity by hinge glycans. Nat. Commun. 14, 7175 (2023).
Yang, H. et al. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. D. Biol. Crystallogr. 60, 1833–1839 (2004).
wwPDB Consortium. EMDB-the Electron Microscopy Data Bank. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad1019 (2023).
Westbrook, J. D. et al. The Chemical Component Dictionary: complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the Protein Data Bank. Bioinformatics 31, 1274–1278 (2015).
Gražulis, S. et al. Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration. Nucleic Acids Res. 40, D420–D427 (2012).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D. Biol. Crystallogr. 65, 1074–1080 (2009).
Nicholls, R. A. et al. The missing link: covalent linkages in structural models. Acta Crystallogr. D. Struct. Biol. 77, 727–745 (2021).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D. Biol. Crystallogr. 60, 2184–2195 (2004).
Chojnowski, G., Sobolev, E., Heuser, P. & Lamzin, V. S. The accuracy of protein models automatically built into cryo-EM maps with ARP/wARP. Acta Crystallogr. D. Struct. Biol. 77, 142–150 (2021).
Terashi, G. & Kihara, D. De novo main-chain modeling for EM maps using MAINMAST. Nat. Commun. 9, 1618 (2018).
Terashi, G., Kagaya, Y. & Kihara, D. MAINMASTseg: automated map segmentation method for cryo-EM density maps with symmetry. J. Chem. Inf. Model. 60, 2634–2643 (2020).
Chen, M. & Baker, M. L. Automation and assessment of de novo modeling with pathwalking in near atomic resolution cryoEM density maps. J. Struct. Biol. 204, 555–563 (2018).
DiMaio, F., Tyka, M. D., Baker, M. L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using rosetta. J. Mol. Biol. 392, 181–190 (2009).
Webb, B. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 1137, 1–15 (2014).
Si, D. et al. Deep learning to predict protein backbone structure from high-resolution cryo-EM density maps. Sci. Rep. 10, 4282 (2020).
Pfab, J., Phan, N. M. & Si, D. DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl Acad. Sci. USA 118, e2017525118 (2021).
Igaev, M., Kutzner, C., Bock, L. V., Vaiana, A. C. & Grubmüller, H. Automated cryo-EM structure refinement using correlation-driven molecular dynamics. eLife 8, e43542 (2019).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D. Biol. Crystallogr. 71, 136–153 (2015).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D. Struct. Biol. 77, 1282–1291 (2021).
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D. Biol. Crystallogr. 70, 2487–2499 (2014).
Singharoy, A. et al. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. eLife 5, e16105 (2016).
Shekhar, M. et al. CryoFold: determining protein structures and data-guided ensembles from cryo-EM density maps. Matter 4, 3195–3216 (2021).
Chang, L., Mondal, A., MacCallum, J. L. & Perez, A. CryoFold 2.0: cryo-EM structure determination with MELD. J. Phys. Chem. A 127, 3906–3913 (2023).
MacCallum, J. L., Perez, A. & Dill, K. A. Determining protein structures by combining semireliable data with atomistic physical models by Bayesian inference. Proc. Natl Acad. Sci. USA 112, 6985–6990 (2015).
Perez, A., MacCallum, J. L. & Dill, K. A. Accelerating molecular simulations of proteins using Bayesian inference on weak information. Proc. Natl Acad. Sci. USA 112, 11846–11851 (2015).
Chojnowski, G. DoubleHelix: nucleic acid sequence identification, assignment and validation tool for cryo-EM and crystal structure models. Nucleic Acids Res. 51, 8255–8269 (2023).
Hsin, J., Arkhipov, A., Yin, Y., Stone, J. E. & Schulten, K. Using VMD: an introductory tutorial. Curr. Protoc. Bioinforma. Chapter 5, Unit 5.7 (2008).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Warshamanage, R., Yamashita, K. & Murshudov, G. N. EMDA: a Python package for electron microscopy data analysis. J. Struct. Biol. 214, 107826 (2022).
Burnley, T., Palmer, C. M. & Winn, M. Recent developments in the CCP-EM software suite. Acta Crystallogr. D. Struct. Biol. 73, 469–477 (2017).
Ramlaul, K., Palmer, C. M. & Aylett, C. H. S. A local agreement filtering algorithm for transmission EM reconstructions. J. Struct. Biol. 205, 30–40 (2019).
Olechnovič, K. & Venclovas, Č. Contact area-based structural analysis of proteins and their complexes using CAD-score. Methods Mol. Biol. 2112, 75–90 (2020).
McDonald, I. K. & Thornton, J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994).
Zemla, A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 31, 3370–3374 (2003).
Mukherjee, S. & Zhang, Y. MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 37, e83 (2009).
Biasini, M. et al. OpenStructure: an integrated software framework for computational structural biology. Acta Crystallogr. D. Biol. Crystallogr. 69, 701–709 (2013).
Chen, V. B., Davis, I. W. & Richardson, D. C. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 18, 2403–2409 (2009).
Rose, Y. et al. RCSB Protein Data Bank: architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB Archive. J. Mol. Biol. 433, 166704 (2021).
Lawson, C. L. et al. 2021 EMDataResource Ligand Model Challenge dataset. Zenodo https://doi.org/10.5281/zenodo.10551958 (2024).
Burley, S. K. et al. Electron microscopy holdings of the Protein Data Bank: the impact of the resolution revolution, new validation tools, and implications for the future. Biophys. Rev 14, 1281–1301 (2022).
Acknowledgements
EMDataResource (C.L.L., A.K., G.D.P., H.M.B., W.C.) was supported by the US National Institutes of Health/National Institute of General Medical Science (NIH/NIGMS) grant no. R01GM079429. The following additional grants are acknowledged for participant support. American Leprosy Missions grant no. G88726 to S.C.V. Biotechnology and Biological Sciences Research Council grant no. BB/S005099/1 to P.S.B. and K.D.C. Biotechnology and Biological Sciences Research Council grant no. BB/T012935/1 to S.W.H. Biotechnology and Biological Sciences Research Council grant no. BB/S007083/1 to R.A.N. German Research Foundation grant no. CIBSS – EXC-2189 – 390939984 to C.H. and W.-C.K. Institute of Biotechnology Czech Academy of Sciences grant no. RVO 86652036 to J. Černý and B.S. Max Planck Society and German Research Foundation grant no. RTG 2756 to H.G. and M.I. Medical Research Council grant no. MR/V000403/1 to C.M.P., T.B., A.P.J. and M.D.W. Medical Research Council grant no. MC_UP_A025_1012 to G.N.M., K.Y. and P.E. NIH/NIGMS grant no. P01GM063210 to M.L.B. and C.H. NIH/NIGMS grant nos. P01GM063210, R01GM073919 and R35GM131883 to J.S.R., M.G.P., C.J.W., V.B.C. and D. C. Richardson. NIH/NIGMS grant nos. P01GM063210, R01GM071939 and R24GM141254. US Department of Energy grant no. DE-AC02-05CH11231 to the Phenix Industrial Consortium: N.W.M., P.V.A., C.J.S. and O.V.S. NIH/NIGMS grant no. R01GM133840, National Science Foundation grant no. IIS2211598 to D. Kihara. NIH/NIGMS grant no. R01GM146340 to J. Cheng and N.G. NIH/NIGMS grant no. R01GM123089 F.D. and A. Muenks. National Science Foundation grant no. DGE-1762114 to A. Muenks. National Science Foundation grant no. CHE-2235785 to A.P. National Science Foundation grant no. DBI-1832184, US Department of Energy grant nos. DESC0019749 and NIH/NCI, NIAID, NIGMS R01GM133198 to S.K.B., C.S. and C.L.L. NSERC of Canada, grant no. RGPIN-05795-2016 to C.N.R. SERB grant no. CRG/2022/002761 to S.M. US Department of Energy grant no. DE-AC02-06CH11357 to A.J. UWB Scholarship, Research and Creative Practice grant no. 2023-2024 to D. Si. Wellcome Trust grant no. 209407/Z/17/Z to R.J.R. Wellcome Trust, grant no. 208398/Z/17/Z to R.W. Funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
H.M.B. and W.C. conceived the project. C.L.L. and A.K. organized the Challenge with the assessors and modelers. G.D.P. and M.F.S. assisted in the analysis. J. Černý, P.E., A.J., J.S.R., R.J.R., A.L.R. and B.S. helped to develop Challenge goals and guidelines. Authors listed in Table 1 prepared and submitted models for the Challenge. Authors listed in Table 2 provided assessment results. C.L.L., A.K., G.D.P., M.S., H.M.B. and W.C. wrote the initial draft. All coauthors participated in review and revision of the paper.
Corresponding authors
Ethics declarations
Competing interests
S.N., A.G., A.R., B.S. and Y.Y. are current or former employees of Genentech. E.S. and C.I.W. are current employees of Chemical Computing Group. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Arunima Singh, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
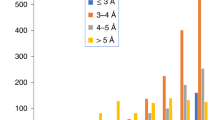
Extended Data Fig. 1 Growth of cryo-EM structures and novel ligands derived from them.
(a) Cryo-EM maps released into the EM Data Bank (EMDB) archive by year and resolution range (source: www.emdataresource.org) up to the end of 2023. (b) Novel non-polymer ligands included in cryo-EM structures by year of release into the Protein Data Bank (PDB) through 2023. Inset: major categories of novel ligands found in cryo-EM-derived models (through 2021). See Online Methods for details.
Extended Data Fig. 2 Selected submitted ligand models for each of the Challenge targets.
Panels are labeled by team ID and model # (see Table 1), in order of decreasing ligand Q-scores (see Fig. 3, row 1) from top to bottom. The portion of the map corresponding to the ligand is shown as a semi-transparent surface, along with the model of the ligand. Ligand Q-score is the average Q-score of all non-H atoms in the ligand. For each atom, the Q-score is measured by correlation of map density to the expected gaussian function, at points within 2 Å of the atom and closer to the atom than any other non-H atom in the the model10. Higher-scoring ligand models fit better in the cryo-EM density than lower-scoring models.
Extended Data Fig. 3 Evaluation of ions in submitted models (stereo images).
(a) Target 1 6cvm reference model Mg A2002 (gray sphere) with water ligands (orange spheres), located near the PETG ligand, with density for classic octahedral coordination. Only six of 23 submitted Target 1 models included the Mg2+ and all three coordinating waters. Others had either only Mg2+, Mg2+ plus one or two waters, Mg2+ plus waters with zero occupancy, no atoms modeled, or atoms significantly displaced. (b) Some groups placed metal ions with weak justification, as exemplified by the Na+ (grey sphere) shown here in model EM005_1 for Target 3.
Extended Data Fig. 4 Q-score rankings for ligand + extended vicinity and for full models.
(a-c) LIVQ10 (Ligand + extended vicinity ≤10 Å) Q-scores (black bars) and full model Q-scores (gray bars) are plotted for each submitted model and each reference model, with order according to ligand + extended vicinity rank.
Extended Data Fig. 5 Alternative Group Ranking by sum of Ligand, Ligand+Environment, Full Model Coordinates-only, Full Model Fit-to-Map composite scores.
(a) Group ranking (left-to-right) according to the sum of four composite z-scores, as described below. Only groups that submitted models for all 3 targets and have rank similar to or better than PDB reference models are shown. (b) Correlation table (n = 61) of scores used to create z-scores and rankings in panel (A) and/or Fig. 4. Group composite scores were calculated per team as follows. For each submitted model, and for each score type, a composite z-score was calculated. For each target (T1, T2, T3), the model submitted by that group with maximum composite z-score was selected for inclusion in the final average score over all targets. Ligand: z = (0.33*z.MogulComposite + 0.33*z.StrainEnergyMM + 0.33*z.Q-ligand). Ligand+environment: z = (0.33*z.Pharmacore + 0.33*z.Probescore + 0.33*z.LIVQ5). Full model coordinates-only: z = (0.25*z.Clash + 0.5*z.CablamConf + 0.25*z.CablamCa). Full model fit-to-map: z = (0.25*z.EMRinger + 0.25*z.Q-Protein + 0.25*z.TEMPySMOC + 0.25*z.PhenixFCS05).
Extended Data Fig. 6 Ligand/Ligand Environment Probescores.
(a) Molprobity Probescore32 distributions for ligands in Targets 1–3 (reference models: red triangles; submitted model scores are plotted as gray circles with following exceptions: Target 1, yellow boxes if PTQ sugar ring position was flipped relative to reference; Target 2, asterisk if F86 was set to half-occupancy; Target 3, blue diamonds if PEE was modeled as head-group+tails). Scores are plotted in horizontal axis lanes with small random vertical shifts to visually separate clustered points. Notably, score distributions have wide spreads independent of noted model features: PTQ sugar orientation, F86 occupancy, or PEE inclusion of tails–although for PEE the score distribution is noticeably broader when the larger and more variable tails are included. (b) T2 density map with reference model in the region of the F86 ligand, showing half-strength density for the remdesivir ligand, implying that only half the molecules have covalently bound inhibitor. Image is reproduced from Figure 6 of reference 38 (open access CC-BY license, no permission required for reuse). (c-e) T2 F86 + pyrophosphate ligand environments for the reference model (PDBid 7BV2), model EM004_2, and model EM008_1, respectively. All-atom contact dots are from Probescore, with all-atom clashes in hot pink and favorable H-bonds and vdW contacts in green and blue. Molecular graphics are shown in KiNG88.
Supplementary information
Supplementary Information
Challenge summary statistics (pp. 1–2), Submission form screenshots (pp. 3–7) and Pharmacore assessment report (pp. 8–18).
Supplementary Data 1
Collected metadata information describing the workflow for each model submitted to the Ligand Challenge.
Supplementary Data 2
Collected assessments, scores and score correlations for models submitted to the Ligand Challenge.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lawson, C.L., Kryshtafovych, A., Pintilie, G.D. et al. Outcomes of the EMDataResource cryo-EM Ligand Modeling Challenge. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02321-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41592-024-02321-7
- Springer Nature America, Inc.