Abstract
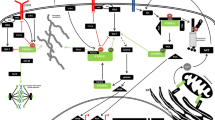
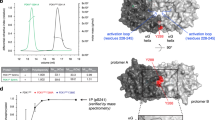
We have identified a molecular interaction between the reversibly oxidized form of protein tyrosine phosphatase 1B (PTP1B) and 14-3-3ζ that regulates PTP1B activity. Destabilizing the transient interaction between 14-3-3ζ and PTP1B prevented PTP1B inactivation by reactive oxygen species and decreased epidermal growth factor receptor phosphorylation. Our data suggest that destabilizing the interaction between 14-3-3ζ and the reversibly oxidized and inactive form of PTP1B may establish a path to PTP1B activation in cells.


Similar content being viewed by others
References
Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7–15 (2011).
Ostman, A., Frijhoff, J., Sandin, A. & Böhmer, F. D. Regulation of protein tyrosine phosphatases by reversible oxidation. J. Biochem. 150, 345–356 (2011).
Tonks, N. K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846 (2006).
Karisch, R. et al. Global proteomic assessment of the classical protein-tyrosine phosphatome and “Redoxome”. Cell 146, 826–840 (2011).
Brown, D. I. & Griendling, K. K. Regulation of Signal Transduction by Reactive Oxygen Species in the Cardiovascular System. Circ. Res. 116, 531–549 (2015).
Tonks, N. K. PTP1B: from the sidelines to the front lines! FEBS Lett. 546, 140–148 (2003).
Feldhammer, M., Uetani, N., Miranda-Saavedra, D. & Tremblay, M. L. PTP1B: a simple enzyme for a complex world. Crit. Rev. Biochem. Mol. Biol. 48, 430–445 (2013).
Salmeen, A. et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 (2003).
van Montfort, R. L., Congreve, M., Tisi, D., Carr, R. & Jhoti, H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 (2003).
Haque, A., Andersen, J. N., Salmeen, A., Barford, D. & Tonks, N. K. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell 147, 185–198 (2011).
Krishnan, N. et al. Harnessing insulin- and leptin-induced oxidation of PTP1B for therapeutic development. Nat. Commun. 9, 283 (2018).
Zhao, J., Meyerkord, C. L., Du, Y., Khuri, F. R. & Fu, H. 14-3-3 proteins as potential therapeutic targets. Semin. Cell Dev. Biol. 22, 705–712 (2011).
Reinhardt, H. C. & Yaffe, M. B. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14, 563–580 (2013).
Andersen, J. N. et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell Biol. 21, 7117–7136 (2001).
Lee, S. R., Kwon, K. S., Kim, S. R. & Rhee, S. G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273, 15366–15372 (1998).
Boivin, B., Zhang, S., Arbiser, J. L., Zhang, Z. Y. & Tonks, N. K. A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proc. Natl Acad. Sci. USA 105, 9959–9964 (2008).
Boivin, B., Yang, M. & Tonks, N. K. Targeting the reversibly oxidized protein tyrosine phosphatase superfamily. Sci. Signal. 3, pl2 (2010).
Wang, B. et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 (1999).
Ravichandran, L. V., Chen, H., Li, Y. & Quon, M. J. Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor. Mol. Endocrinol. 15, 1768–1780 (2001).
Yang, L. et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 64, 4394–4399 (2004).
Milarski, K. L. et al. Sequence specificity in recognition of the epidermal growth factor receptor by protein tyrosine phosphatase 1B. J. Biol. Chem. 268, 23634–23639 (1993).
Biscardi, J. S. et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 274, 8335–8343 (1999).
Lessard, L., Stuible, M. & Tremblay, M. L. The two faces of PTP1B in cancer. Biochim. Biophys. Acta 1804, 613–619 (2010).
Dagnell, M. et al. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-β receptor tyrosine kinase signaling. Proc. Natl Acad. Sci. USA 110, 13398–13403 (2013).
Parsons, Z. D. & Gates, K. S. Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry 52, 6412–6423 (2013).
Zougman, A., Selby, P. J. & Banks, R. E. Suspension trap** (STrap) sample preparation method for bottom‐up proteomics analysis. Proteomics 14, 1006–11010 (2014).
Ross, P. L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 3, 1154–1169 (2004).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Acknowledgements
We thank H. Fu for providing the 14-3-3ζ plasmid. This research was supported by the National Institutes of Health grant no. HL138605 and the American Heart Association grant no. 17GRNT33700265 to B.B. and National Institutes of Health grant no. GM55989 to N.K.T. B.B. is also grateful for support from the following foundations: the Heart and Stroke Foundation of Canada and SUNY Research Foundation. B.B. is an FRQS Research Scholar and A.B. was the recipient of a scholarship from the FRQS.
Author information
Authors and Affiliations
Contributions
A.D.L., A.B., S.M.C., A.K., G.C., S.H.M.R. and B.B. performed experiments and analyzed data. K.D.R. and D.J.P. acquired and analyzed MS data. S.J.K. performed structural analysis and modeling. F.Z. and R.J.L. acquired and analyzed SPR data. N.K.T. and B.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Figs. 1–21.
Rights and permissions
About this article
Cite this article
Londhe, A.D., Bergeron, A., Curley, S.M. et al. Regulation of PTP1B activation through disruption of redox-complex formation. Nat Chem Biol 16, 122–125 (2020). https://doi.org/10.1038/s41589-019-0433-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0433-0
- Springer Nature America, Inc.
This article is cited by
-
14-3-3ζ inhibits maladaptive repair in renal tubules by regulating YAP and reduces renal interstitial fibrosis
Acta Pharmacologica Sinica (2023)
-
Protein disulfide isomerase A1 as a novel redox sensor in VEGFR2 signaling and angiogenesis
Angiogenesis (2023)
-
Reactive oxygen species (ROS) as pleiotropic physiological signalling agents
Nature Reviews Molecular Cell Biology (2020)