Abstract
Wearable sweat sensors with various sensing systems can provide noninvasive medical diagnostics and healthcare monitoring. Here, we demonstrate a wearable microfluidic nanoplasmonic sensor capable of refreshable and portable recognition fingerprint information of targeted biomarkers including urea, lactate, and pH in sweat. A miniature, thin plasmonic metasurface with homogeneous mushroom-shaped hot spots and high surface-enhanced Raman scattering (SERS) activity is designed and integrated into a microfluidics platform. Compared to conventional wearable SERS platforms with the risk of mixed effect between new and old sweat, the microfluidic SERS system allows sweat administration in a controllable and high temporal-resolution fashion, providing refreshable SERS analysis. We use a portable and customized Raman analyzer with a friendly human-machine interface for portable recognition of the spectroscopic signatures of sweat biomarkers. This study integrates epidermal microfluidics with portable SERS molecular recognition, presenting a controllable, handy, and dynamical biofluid sensing system for personalized medicine.
Similar content being viewed by others
Introduction
Flexible smart electronics have revolutionized our cognition, methodology, and techniques in electronic skin, human-machine interaction, personalized healthcare1,2,3,4,5,6,7,8,9. More specifically, wearable sweat sensors that enable cognition of molecular-level signatures related to physiological information in epidermally available sweat are regarded as an extremely competitive device10,11,12,13. Remarkable progress has been made in these wearable sweat sensors by combining molecular recognition methods, micro-nano device manufactures, integrated hardware/software systems, and various analytical techniques14,15,16. Wearable colorimetric27,28. Electrochemical techniques are featured with high sensitivity and selectivity, where the electrodes can also be flexibly designed into different shapes and sizes. Each sensing strategy has its own merits and shortcomings (Supplementary Table 1). Continuous innovations towards develo** new signal reading techniques are necessary to furnish our options for designing wearable sweat sensors.
SERS is a commonly used analytical technique capable of achieving high enhancement of Raman signals through localized plasmon-enhanced excitation and scattering29,30,31. Flexible plasmonic devices by integration of SERS with wearable techniques have attracted tremendous attention for diverse wearable biomedical applications32,33,34,35,36,37. At the present, only few wearable SERS sweat sensors have been demonstrated35,38,39. However, these non-microfluidic sweat systems rely on sweat-permeable (porous) SERS substrates that allow sweat to wick and occupy the hotspots. However, the cost is that these permeable SERS substrates are usually structurally unstable and vulnerable to epidermal deformation when touching the skin. By comparison, microfluidics can spatially control the localization of SERS substrates, and the SERS substrates can be flexibly chosen and configured. Additionally, dynamical sweat transporting enabled by microfluidics can minimize the mixing and carry-over effect of the new and old sweat40, ensuring the SERS analysis is performed in a refreshable and high temporal resolution manner. Another significant issue of the previously-proposed wearable sweat SERS platforms is the readout system. The conventional heavy Raman instrument limits wearable SERS analysis in standardized laboratory settings, which dramatically weakens the practicality and applicable circumstances.
In this paper, we propose a wearable plasmonic sensor, bridging the gap between wearable sweat microfluidics with the SERS signal output technique using a portable Raman analyzer. As shown in Fig. 1, the polydimethylsiloxane (PDMS) microfluidics achieves sweat collection with programmable flowing routing and eliminates contamination and evaporation. In our microfluidic system, the used SERS substrate does not need to play the role of sweat sampling. Thus, an intrinsically connected SERS substrate with fine structural entirety is chosen and independently embedded in the well-defined vessel of microfluidics for in situ and continuous SERS sensing. And compared to the recently proposed paper-based microfluidic SERS device41, this PDMS-based microfluidic can provide a volumetric microchamber for better fixing the SERS substrate. The portable SERS analyzer (commercial device, not customized one) can decode the sweat fingerprint information of targeted biomarkers of urea, lactate, and pH at the molecular level, significantly expanding the accessible scenarios of wearable SERS sensors for point-of-care test (POCT) applications.
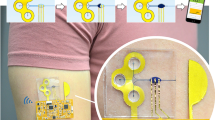
a Stacked view showing each layer of the integrated device. b Schematic of all-in-one biofluid flowing, store, and SERS analysis based on a microfluidic plasmonic device. c Actual image of an as-prepared microfluidic SERS sensor attached to the skin. d System-level block diagram showing the internal functional modules of the portable Raman analyzer.
Results
Design of the wearable microfluidic sweat SERS sensor
The integrated microfluidic SERS platform contains four layers from bottom to up (Fig. 1a): (i) double-sided medical adhesive tape as an epidermal adhesive layer; (ii) microfluidic PDMS layer; (iii) two ultrathin SERS chips inside the microchamber of the microfluidic layer; (iv) a Kapton tape encapsulation layer. The biofluid can flow into the microchannels driven by the pressure action of the subcutaneous sweat glands and assisted by capillary effect in the microchannels. When sweat finally occupies the plasmonic metasurface, in-situ Raman analysis is operable relying on the SERS effect generated by the ordered silver metasurface. Thus, molecular signatures of targeted analytes in sweat can be extracted (Fig. 1b). An actual photograph of the integrated sensor mounted on one volunteer’s back is shown in Fig. 1c, where the inlets, outlet, microchannels, microchamber, and SERS chips can be clearly observed. The whole structural layout is so concise that no complex circuit, battery, or conductive routes are required, which will otherwise bring an extra structural burden and weaken the wearability of the whole device. In this study, the main SERS analytical work is performed using a portable Raman spectrometer, whose optical image is shown in Fig. 1d. The portable Raman device consists of six main modules: core (control unit), laser driver (laser emitting), CCD (charge coupled device), peripheral circuit (signal transmission), Raman module (signal analysis), and smart-port (software section). In contrast to the conventional heavy desktop Raman spectrometer, such a portable Raman analyzer is so convenient that allows users to track the sweat fingerprint as free as they can.
Characterizations of the Ag nanomushroom arrays
Figure 2 illustrates the comprehensive characterizations of our SERS substrate. Vertically aligned noble metal (Ag) nanostructures were selected here for generating local surface plasmon resonance (LSPR) effect according to electromagnetic enhancement theory42. Based on template-assisted reactive ion etching (RIE) techniques (Supplementary Fig. 1), we first fabricated Si nanopillar arrays (Fig. 2a). The high-activity SERS substrate was obtained by further sputtering a layer of Ag on Si nanopillar arrays. This method combines low-cost etching and large-area sputtering techniques, which can potentially bridge the gap between fundamental research and large-scale commercialization. The peaks at Ag 3d5/2 (368.3 eV) and Ag 3d3/2 (374.3 eV) in Supplementary Fig. 2a and Supplementary Fig. 2b indicate the successful introduction of metallic Ag. Energy dispersive X-Ray (EDX) spectroscopy further suggests that the Ag element was uniformly covered on Si nanopillar arrays (Supplementary Fig. 2c). The resulting metasurface was composed of dense, rough, and hierarchically mushroom-shaped nanostructures. Each nanomushroom architecture was 120-130 nm in diameter and 180-200 nm in height (Fig. 2b and Supplementary Fig. 3a), and the gap was estimated to be less than 10 nm between each other. The AFM image in Fig. 2c show the details about the top surface of Ag nanomushroom, consisting of rough and irregular convex aggregate with a diameter ranging from 15 to 45 nm.
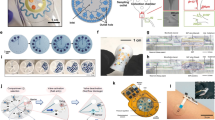
a Scanning electron microscope (SEM) image of Si nanopillar arrays fabricated by template-assisted ion etching techniques. b SEM and c atomic force microscope (AFM) images of Ag nanomushroom arrays after deposition of Ag layer on Si nanopillar. d FEA and the e enlarged view of local electric field distribution over plasmonic nanogaps among Ag nanomushroom arrays. f Comparison of Raman spectra and g corresponding AEFs between Ag layer deposited on bare and Si nanopillar arrays. h Initial repeatability assessment of Raman intensity of R6G collected from 30 random sites of the Ag nanomushroom arrays. i Raman intensity map** of the Ag nanomushroom arrays. Scale bar: 5 μm. Photographs of the as-prepared flexible microfluidic SERS patch under j bending and k pressing measurements, and l the corresponding intensity variations of Raman signals. The R6G concentrations were 10-8 M in h, i, and l, and all the intensity values were extracted from the characteristic band at ~1364 cm−1. The error bars represent standard deviations from three different samples.
The finite element analysis (FEA) was performed to show the electromagnetic field distribution of the Ag nanomushroom arrays. The simulation results confirmed that the hot spots region (yellow and orange color) was highly distributed in the gaps (set as 5 nm) between each Ag nanomushroom, as illustrated in Fig. 2d and e (785 nm), with maximum electromagnetic field intensity (Ex) of ~10 V/m. Similar hot spot distributions were also found in the case of applying 633 and 532 nm plane wave illumination beams, except for a slight decrease in the Ex (Supplementary Fig. 4). Thus, positioning targeted molecules within these nanogap sites can produce a fine electromagnetic enhancement effect, which was further confirmed by subsequent experiment results. The Raman peaks of 10−3 M rhodamine6g (R6G) molecules on Si substrate (dried state) were difficult to observe (Fig. 2f). After supering a layer of Ag on a Si substrate, the Raman intensity improved slightly but was still quite weak, with an analytical enhancement factor (AEF) calculated to be 1.68 (Fig. 2g). The AEF was calculated according to the following equation: AEF = (ISERS/CSERS)/(IRS/CRS), where CSERS and CRS denote the concentrations of R6G on the targeted SERS substrate (here, planar Ag and nano Ag substrate) and reference substrate (here, Si substrate), respectively, ISERS and IRS indicate the corresponding Raman intensity. By comparison, the peak intensity of 10−8 M R6G in Ag nanomushroom arrays was significantly stronger than the above two ones, with AEF up to 1.488 × 106. Additionally, bare Ag nanomushroom arrays without R6G do not produce any Raman peak. Repeatability is another key performance for reliable SERS detection. We first randomly extracted 30 SERS spectra from the R6G-modified Ag nanomushroom arrays (Fig. 2h), whose intensity was roughly comparable with the relative standard deviation (RSD) low to 5.9%. SERS map** was further performed to evaluate the spot-to-spot consistency of the plasmonic substrate. Figure 2i demonstrates that the variation was less than ±10%, exhibiting favorable signal reproducibility and homogeneity.
The integrated SERS microfluidics was achieved by fixing the obtained Ag nanostructures inside the microchamber of the microfluidic layer, followed by encapsulation using double-sided tape (bottom layer) and Kapton tape (top layer, Supplementary Fig. 5a). Kapton tape was also transparent (Supplementary Fig. 5b), and the laser could pass through it (Supplementary Fig. 5c). Therefore, the obtained characteristic peaks of R6G were comparable to those of not using Kapton tape, except for few losses in signal intensity as shown in Supplementary Fig. 5d. More importantly, Kapton tape was chosen here due to its fine thermal stability that could minimize the potential unfavorable heat effect (wrinkling or invalid encapsulation of the microfluidic) caused by laser. Although the SERS substrate consisting of Ag-deposited Si nanopillar arrays was rigid, given its ultrathinness (only 100 μm in thickness) and miniaturization (3.9 mm in diameter), as illustrated in Supplementary Fig. 3b and Supplementary Fig. 3c, the integrated SERS microfluidics was still flexible and maintained stable SERS signal regardless of ex-vivo or on-body mechanical stimulations (Fig. 2j–l).
Spectral calibrations of the microfluidic SERS system
The SERS sensing performance of each sensor was experimentally determined with standard solutions containing different targeted analytes using a portable Raman analyzer. Urea and lactate assays were performed in a label-free manner (Fig. 3a), where no modifications to the SERS substrate were required, greatly simplifying the analytical procedures. Their SERS spectra are shown in Fig. 3b and c. (see Supplementary Table 2 for assignment for each peak)43. The sharp dominant peaks at 1005 and 853 cm−1 were employed for calibration of urea and lactate, and linear standardized curves of urea from 0.1 to 1000 mM, lactate from 0.01 to 100 mM were obtained with correlation coefficients of 0.999 and 0.950, respectively (Supplementary Fig. 6a and Supplementary Fig. 6b). For pH sensing as indicated in Fig. 3d, pH-sensitive probe 4-mercaptobenzoic acid (4-MBA) were pre-modified on the Ag nanomushroom arrays based on Ag-S bonding, which protonated at an acidic pH and deprotonated at a basic pH, respectively. As pH increased from 4 to 8, the Raman band between 1400 and 1425 cm−1 (attributed to carboxylate stretching37, referred to cbx hereinafter) underwent an obvious blue shift (Fig. 3e). After normalizing the Raman intensity at the 1078 cm−1 band (aromatic ring vibrations), the ratio of Icbx/I1078 could be used to calibrate pH from 4 to 7 with a correlation coefficient of 0.947 (Fig. 3f and Supplementary Fig. 6c).
a Schematic and SERS spectra of b urea and c lactate in a label-free manner. d The mechanism of pH SERS sensing showing the probe molecule of 4-MBA labeled on the Ag surface, which can be protonated at an acidic pH and deprotonated at a basic pH. e SERS spectra and f the enlarged view of the Raman peak at 1400-1425 cm−1 responded to different pH levels after normalizing the peak intensity at 1078 cm−1. g Schematic of simultaneous and multiple SERS analysis of sweat targets based on one microfluidic chip. h Compatibility study of simultaneous SERS detection of urea and lactate. i Interference study of pH SERS sensing.
In our integrated microfluidic SERS system, the SERS analysis in label-free and label manner were respectively configurated in two independent microchambers (Fig. 3g). Simultaneous SERS analysis of urea and lactate could be performed in a label-free manner. As illustrated in Fig. 3h, most Raman peaks were visually perceptible in both individual urea or lactate solutions as well as their mixtures. For pH assay, as the Ag nanomushroom arrays were prelabeled with the pH-responsive 4-MBA molecules, the functionalized hotspots were inactive to other analytes (e.g., urea and lactate). Therefore, fine analytical selectivity was found, and negligible cross influence on the Raman signal was observed (Fig. 3i).
Dynamic sweat sampling and analysis performances
One of the advantages of this microfluidic SERS system is its individual design of sweat inlets and SERS analytical site. In conventional non-microfluidic wearable SERS sensing systems, SERS substrates are directly in contact with the skin (Fig. 4a). In that case, these SERS substrates were usually designed to be sweat-permeable (e.g., nanocube35, nanowires38) so that sweat could be wicked to occupy the hotspots. However, permeable SERS substrates tend to produce defects, especially during intensive exercise (e.g., stretching, twisting), which increases the potential risk when a laser passed through these defects (in micron-scale) and to the skin below. Here we use microfluidics instead, where sweat could be introduced from individual inlets. Thus, non-permeable SERS substrates (here, Ag nanomushroom arrays on Si wafer) with fine structural entirety can be used. In this way, even if there are major cracks in Ag nanomushroom arrays, the laser can be completely blocked against skin by the Si wafer (Fig. 4b), without the need of using a laser block component41.
a, b Schematic illustrating the comparison between non-microfluidics & permeable SERS substrate and microfluidics & non-permeable SERS substrate system. c Photographic images of a complete sweat sampling process based on the microfluidic patch. d FEA of the solute concentration distributions over the microchamber at different time instances. e Schematic, f Raman spectrogram, and g intensity analysis (at 853 cm−1) of 12 SERS measurement cycles of lactate showing the reversibility of the sensor for dynamic sweat analysis. h Schematic, i Raman spectrogram, and j standardized Icbx/I1078 of pH sensing in different solution volumes showing the low-volume analysis in the microfluidic SERS platform. The error bars represent standard deviations from three different samples.
Another advantage enabled by epidermal microfluidics is the dynamical SERS analysis with sufficient spatiotemporal resolution. We investigated the relationship between sweat dynamics and SERS sensing performance based on this microfluidic SERS system. The sweat flow rate was first measured. The on-body sweat sampling experiment in Fig. 4c reveals that the freshly secreted perspiration stepwise flowed in the streamline channels and occupied the central microchamber. Based on the whole empty volume (~11.8 μL) and biofluid filling time (~14 min), the total flow rate was calculated to be about 0.84 μL/min, viz., 0.14 µL/min for each inlet. The time required for a solute concentration in the reservoir to switch to the new level when introducing fresh inflow (also named refreshing time in some literature44), is an important index for continuous biofluid monitoring. We analyzed the refreshing time over the microchamber (the volume of the SERS chip was deducted) by FEA, using lactate as model solute with the above calculated flowing rate. Figure 4d reveals that as the lactate changed from 1 mM to 10 mM, the refreshing time taken to reach 90% of the new solute concentration was about 8 min. Not surprisingly, increasing the inlet numbers from 2, 4, 6, 8 to 10 led to a faster refreshing behavior (Supplementary Fig. 7a and Supplementary Fig. 7b). However, as shown in Supplementary Fig. 7c, the increase rate became obviously slow when the inlets exceeded 6. That is why the inlet number was finally optimized to 6 in our work.
Based on this refreshable microfluidic system, dynamic SERS analysis was conducted in Fig. 4e via continuous analyte solution injection that mimicked the human sweating process. Cycling the lactate solutions between 1 to 10 mM at an interval of 8 min (based on the above simulated refreshing time) also resulted in a synchronous cycle of Raman signals, without obvious intensity loss (less than 1%, Fig. 4f and g). The inset in (f) indicates the step changes in lactate concentration experienced by the sensor. A similar phenomenon was also observed in cyclic urea and pH measurements (Supplementary Fig. 8), displaying reversible and continuous SERS sensing performance. Such a microfluidic device ensures continuous freshly-secreted sweat sampling. In this way, the risk of mixed effect between new and old sweat can be minimized10, which is a key improvement compared to non-microfluidic sweat SERS system35,38,39. Surprisingly, only 0.5 μL of sample solution was sufficient to produce Raman signal for pH measurement. When altering the sample volume from 0.5 to 4 μL, the signal intensity was almost consistent as demonstrated in Fig. 4h–j. These encouraging results indicate that the wearable SERS microfluidic platform is capable of dynamic and ultralow-volume biofluid monitoring.
On-body evaluation of the microfluidic sweat SERS system
Figure 5 depicts the on-body biofluid monitoring enabled by the microfluidic SERS device. As shown in Fig. 5a and Supplementary Fig. 9, a healthy subject was commanded to perform a stationary pulling-down exercise, and the flexible wearable platform can be comfortably patched on various body parts. Biochemical information in the extracted sweat was recorded and screened on a user’s interface of a portable Raman analyzer (Fig. 5b), and the system-level beam path diagram was demonstrated in Fig. 5c (detailed in Methods). In contrast to conventional heavy desktop spectrometers requiring a whole lab room, such a portable analyzer was remarkably miniature and convenient (Supplementary Fig. 10), making the signal output available even at home. The portable device enabled a customizing molecular library (by inputting the spectrum information of standard analytes in advance), thus the measured data can be matched with this pre-built standard library, and transmitted to other devices by Wi-Fi or Bluetooth techniques (Supplementary Fig. 11 and Supplementary Table 3). Figures 5d and e display the discrete spectroscopic signatures in sweat urea, lactate, and pH at specific time points. The characteristic molecular fingerprint of urea at 1005 cm−1 can be found almost throughout the whole exercise. Based on the above calibration plots, we estimated that the urea contents were mainly around 10 mM as shown in Fig. 5f. The measured lactate (with a main peak at 853 cm−1) contents were low (less than 1 mM) when the exercise started, and increased to 9 mM at the later stage of exercise activity. This could be attributed that anaerobic exercise occurred at that time. As reviled by some clinical research, lactate was usually accompanied by anaerobic exercise45. Generally, the measured sweat urea and lactate were slightly lower than some literature values46,47, but were still in a reasonable physiological range10. The standardized pH spectroscopic signatures did not change significantly, and the corresponding pH value was calculated to be 5.5–7.0, indicating that the extracted sweat was weakly acid10. These on-body SERS results had close readings and tendencies with those based on commercial benchmark methods (urea test kit, lactate test kit, and pH meter). Beyond these targets, several characteristic peaks around 1220–1370 cm−1 were also observable in Fig. 5d, which perhaps originated from the C-H deformation and Amide III of proteins in the extracted sweat43.
a Photograph of a volunteer wearing a microfluidic SERS patch during continuous exercise. The inset indicates the portable Raman analyzer. b The user interface and c system-level beam path diagram of the portable Raman analyzer. Discrete Raman spectra of d sweat urea, lactate, and e pH as well as f the corresponding contents calculated from the above calibration curves. The color and black data correspond to measurements done by SERS and by commercial benchmark methods (urea test kit, lactate test kit, and pH meter), respectively. g Schematic showing the metabolic behavior of urea in the human body. Evaluation of the microfluidic SERS device in dietary challenges by comparing the sweat urea and serum urea h with and i without protein intake (n = 6, the points represent raw data; the five lines from bottom to top represent minimum, lower quartile, median, upper quartile and maximum, respectively).
To further assess the feasibility and potential clinical applications of such an epidermal SERS platform, a controlled dietary experiment was performed using urea (with potential correlation with kidney functioning48) as a model marker (Fig. 5g). Briefly, the volunteer’s urea contents in both sweat and blood were measured before/after a standardized protein intake, where the sweat urea was detected based on the above protocol, and blood urea was determined by UV − visible (UV − vis) absorption using a commercial serum urea kit (Supplementary Fig. 12). Figure 5h illustrates that the measured sweat urea 2 hours after protein intake was remarkably higher than that of initial intake. A similar trend in blood urea was also observed, which could be attributed to the migration of urea from the blood vessel to the sweat glands. By comparison, in the case of non-protein intake, both sweat and blood urea abundance showed obvious decreasing trends two hours later (Fig. 5i). Overall, these results demonstrate that our microfluidic SERS was able to noninvasively obtain physiological metabolite information, which can be an alternative strategy for clinical diagnostics.
Discussion
This research has successfully achieved portable SERS sensing of biomarkers in human sweat based on an integrated plasmonic microfluidic patch. This wearable microfluidic biodevice allowed flexible selection and precisely encapsulating highly-active SERS substrate, providing high-temporal-resolution and refreshable sweat SERS analysis in contrast to the non-microfluidic system. The use of a portable Raman analyzer brought convenient access to Raman signal readout anytime and anywhere, avoiding heavy equipment at a given laboratory. The plasmonic microfluidic system enabled rapid, facile, portable, and POCT of physiologically related biomarkers in sweat, providing a clinical research platform for personalized healthcare.
Despite encouraging achievements, there are also some concerns about the engineering of such wearable and portable SERS biofluid sensors (Supplementary Table 1) that should cause our attention. One is the potential peak overlay, especially in the case of multiple label-free SERS analysis. Thus, the rational design of an analytical system after considering the intrinsic peak of high-abundance markers in specific biofluids is necessary. Another issue is repeatability and stability of SERS substrate. A highly uniform substrate regardless of mechanical deformation for repeatable SERS measurement is still urged. The oxidation issue of our Ag nanomushroom SERS substrate is a challenge that should also be overcome in future work. Additionally, the versatility of the portable Raman analyzer in wider scopes of analytes should be further inspected by collecting more experiment data. Still, updated progress has been made in this wearable sweat biosignal output system for non-invasive probing of our bodies at molecular levels. We believe our wearable device will unlock more applications and open up previously unnoticed horizons in various fields, such as athletic, military, criminal investigation, clinical care, intelligent medicine, etc.
Methods
Measurements
The X-ray photoelectron spectrometer of Ag nanomushroom arrays was acquired from Thermo Scientific K-Alpha + , USA. The morphologies of the SERS probe were obtained using SEM (Thermo Scientific APREO S, USA). Raman spectra were mainly recorded on a portable Raman spectrometer (commercial device, not designed by this work, Optosky Photonics Inc, China). For R6G analysis (dry status), the measurement parameters were set as: optical power = 150 mW, integration time = 5 s. For sweat targets analysis (aqueous status), the measurement parameters were set as optical power = 200 mW, integration time = 15 s. Raman map** was obtained by laser scanning Raman microscope (Raman-11, Nanophoton Corporation, Japan). The map** parameters was set as: optical power = 0.83 mW, integration time = 15 s, through a 40×, 0.75 NA objective. Urea and lactate test kits (Wuhan Szybio Co., Ltd, China) were used under assistance of a UV-1800 spectrophotometer (Shimadzu, Japan). Sweat pH was verified by a pH meter (Mettler Toledo, Switzerland).
The detection mechanism of the portable Raman spectrometer is as follows (Fig. 5c): The emitted parallel laser (785 nm) was irradiated through a dichroic filter, then reflected onto the doublet lens (positioned at a 45° angle) and focused on the analytes. After being scattered from the targets and passing through the doublet lens, the laser beam became a collimated beam and was filtered by the dichroic mirror (~95% elastic scattering light of 785 nm is filtered). Then, the Raman signal was unobstructed through the filter group (Semrock) (more than 790 nm transmittance), however, the laser signal OD10 (residual laser 10−10) was filtered. The beam was successively focused by the coupling lens, collimated by a collimator, diffracted by a grating, reflected and focused by the focus mirror, and finally reached the CCD detector for light splitting detection. Thus, the spectral outcomes could be obtained and screened on the user interface.
Fabrication of Ag nanomushroom arrays
Commercial ultrathin Si (n-type, in thickness of 100 μm) wafer was first immersed in piranha solution of mixture H2SO4 (98%): H2O2 (30%) at V/V = 3:1 for 1 h, followed by ultrasonic clean in ethanol (>99.8%), acetone (>99.5%) and ultrapure water (≥18 MΩ, Milli-Q) for 30 min, respectively, and finally blown with nitrogen gas. Then, a monolayer of polystyrene (PS, with diameters of 120 nm was closely packed on the Si wafer by ethanol-assisted self-assembly technique49. The samples were baked at 120 °C for 1 min, making the PS colloidal monolayer closely in contact with the Si wafer. By employing these PS spheres monolayer as a mask, well-aligned Si nanopillar arrays were created by plasma etching in a reactive ion etching machine (ICP-RIE plasma etcher SI-500, Germany, power: 150 w; gas flow rate: O2 20 sccm & SF6 20 sccm; chamber pressure: 2 Pa; etching time: 50 s). After removing residual PS spheres by calcination at 400 °C in a muffle furnace, the Si NP arrays were sputtered with a thin Ag layer using an ion sputtering apparatus at a constant current of 30 mA for 5 min. The SERS chips consisting of Ag nanomushroom arrays were cut into small circular pieces (in diameter of 3.9 mm). Noted that these chips should be kept in vacuum packaging or immersed in ethanol when not used for avoiding oxidation of Ag.
Fabrication of microfluidic layer
PDMS prepolymer (with curing agent at a weight ratio of 10:1. RTV615, USA) was poured onto a SU8 mold (by soft lithographic techniques) to pattern the inlets, reservoir as well as microchannels. The thickness of the PDMS film was controlled to be about 400 μm after curing. The six small cylindrical inlets had a diameter of 1.5 mm, and the large cylindrical collection reservoir possessed a diameter of 4 mm. Both the six inlets and collection reservoir had a depth of 200 μm when peeling from the SU8 mold but then were adjusted to 400 μm using punchers (throughout the PDMS film). All the microchannels were designed to be 350 μm by 200 μm in width and depth, respectively. The PDMS was treated with plasma (180 W for 5 min), as the hydrophilic surface is better for sweat sampling.
SERS assay of targeted markers in standard solutions
All the SERS analyses of standard analytes were performed using 785 nm wavelength under ambient temperature, and all the volumes of analytes were set as 1 μL unless otherwise specified. The R6G, urea, and lactate were directly detected after applying the corresponding solutions to SERS substrate. For pH assay, Ag nanomushroom arrays were incubated in 10 mM 4-MBA (ethanol as solvent) for 45 min and then rinsed with ethanol. Then solutions with different pH were carefully drop-cast onto the 4-MBA-modified plasmonic interface and the remaining SERS detection steps were similar to those above.
FEA of electromagnetic field among the Ag nanomushroom arrays
The FEA of the local electromagnetic field among Ag nanomushroom arrays was performed using COMSOL Multiphysics 5.6. In this calculation, each Ag nanomushroom was simplified as a combined geometry of a circular (head of the Ag nanomushroom), isosceles trapezoid (stem of the Ag nanomushroom), and rectangle (the Ag layer at bottom). The Ag nanoparticles on the head and stem of individual Ag nanomushroom were modeled as inferior arcs. The gap between individual Ag-mushroom arrays was set as 5 nm. Linearly polarized 785, 633, and 532 nm plane wave illumination beams were respectively directed onto the plasmonic nanostructures with polarization along the array axis. The optical parameter of Ag was adopted from previous work50.
FEA of solute diffusion in the microfluidics
The FEA of fluid dynamics involved building a coupled physical field of transport of dilute species transfer and laminar flow using COMSOL Multiphysics 5.6 based on the actual three-dimensional size of our microfluidic system. Before the simulation, we first confirmed the Reynolds number (Re) according to the following equation51:
where ρ, v, and μ represent the density, flow velocity, and viscosity of the liquid, d denotes characteristic length, viz., the width of microchannel in our case. These fluid parameters were set as equivalent to water, as the composition of perspiration is 99% water. According to Eq. (1), Re was calculated to be much lower than 2000 (the critical value of laminar flow). Therefore, the FEA could be conducted by numerically solving the Stokes equation for an incompressible flow coupled with the convection-diffusion equation44:
Where p and C indicate pressure and solute concentration, respectively. D is the solute diffusion coefficients in dilute solutions. For the solute whose molecular weight is less than 1000, D can be roughly calculated as52:
Where α, MB, and T denote the association factor, the molar mass of solvent B, and temperature, respectively. VA (cm3 mol−1) indicates the molecular volume of solute A at a normal boiling point. The average concentration over the central reservoir was computed to track the solute refreshing profile. A standard unit was used for each parameter unless otherwise specified.
Experiments on human subjects
All the experiment protocols on human subjects were strictly performed by the local institutional review board. A healthy male subject aged 31 was recruited and gave written, informed consent. One test zone of the volunteer’s arm was thoroughly washed and swabbed by a medical alcohol strip before placing the sensor patches. Then the participant was recommended to do exercise so that the generated sweat could be continuously collected in the circular reservoir and occupied the inside SERS chips. A protein-rich diet investigation was further carried out by employing urea as a model analyte. In general, we analyzed the urea changes in sweat & serum before (0 h) and after (2 h) protein intake (30 g) as well as non-protein intake. Sweat urea was determined similar to the above experiment protocols, serum urea using serum urea kit based on the urease glutamine-dehydrogenase method) after collecting blood with centrifuge tubes after centrifuging at 6000 rpm for a quarter.
Data availability
All data are available within the article or available from the authors upon reasonable request.
Code availability
There is no code produced in this paper.
References
Dai, Y., Hu, H., Wang, M., Xu, J. & Wang, S. Stretchable transistors and functional circuits for human-integrated electronics. Nat. Electron. 4, 17–29 (2021).
Ray, T. R. et al. Bio-integrated wearable systems: a comprehensive review. Chem. Rev. 119, 5461–5533 (2019).
Gao, W., Ota, H., Kiriya, D., Takei, K. & Javey, A. Flexible electronics toward wearable sensing. Acc. Chem. Res. 52, 523–533 (2019).
Yang, Y. & Gao, W. Wearable and flexible electronics for continuous molecular monitoring. Chem. Soc. Rev. 48, 1465–1491 (2019).
Kim, J., Campbell, A. S., de Ávila, B. E.-F. & Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 37, 389–406 (2019).
Niu, S. et al. A wireless body area sensor network based on stretchable passive tags. Nat. Electron. 2, 361–368 (2019).
Li, L. et al. Ultrastretchable fiber sensor with high sensitivity in whole workable range for wearable electronics and implantable medicine. Adv. Sci. 5, 1800558 (2018).
He, X., Fan, C., Xu, T. & Zhang, X. Biospired janus silk E-textiles with wet-thermal comfort for highly efficient biofluid monitoring. Nano Lett. 21, 8880–8887 (2021).
Su, Y., Lu, L. & Zhou, M. Wearable microbial fuel cells for sustainable self-powered electronic skins. ACS Appl. Mater. Interfaces 14, 8664–8668 (2022).
Bariya, M., Nyein, H. Y. Y. & Javey, A. Wearable sweat sensors. Nat. Electron. 1, 160–171 (2018).
Choi, J., Ghaffari, R., Baker, L. B. & Rogers, J. A. Skin-interfaced systems for sweat collection and analytics. Sci. Adv. 4, eaar3921 (2018).
Heikenfeld, J. et al. Accessing analytes in biofluids for peripheral biochemical monitoring. Nat. Biotechnol. 37, 407–419 (2019).
Sun, M. et al. A flexible and wearable epidermal ethanol biofuel cell for on-body and real-time bioenergy harvesting from human sweat. Nano Energy 86, 106061 (2021).
Ghaffari, R. et al. Soft wearable systems for colorimetric and electrochemical analysis of biofluids. Adv. Funct. Mater. 30, 1907269 (2019).
Bandodkar, A. J., Jeerapan, I. & Wang, J. Wearable chemical sensors: present challenges and future prospects. ACS Sens. 1, 464–482 (2016).
**, X., Liu, C., Xu, T., Su, L. & Zhang, X. Artificial intelligence biosensors: challenges and prospects. Biosens. Bioelectron. 165, 112412 (2020).
**ao, J. et al. Microfluidic chip-based wearable colorimetric sensor for simple and facile detection of sweat glucose. Anal. Chem. 91, 14803–14807 (2019).
Koh, A. et al. A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat. Sci. Transl. Med. 8, 366ra165 (2016).
Bandodkar, A. J. et al. Soft, skin-Interfaced microfluidic systems with passive galvanic stopwatches for precise chronometric sampling of sweat. Adv. Mater. 31, 1902109 (2019).
Xu, X.-Y. & Yan, B. A fluorescent wearable platform for sweat Cl− analysis and logic smart-device fabrication based on color adjustable lanthanide MOFs. J. Mater. Chem. C. 6, 1863–1869 (2018).
Sekine, Y. et al. A fluorometric skin-interfaced microfluidic device and smartphone imaging module for in situ quantitative analysis of sweat chemistry. Lab Chip 18, 2178–2186 (2018).
Zhou, Z. et al. Luminescent wearable biosensors based on gold nanocluster networks for “turn-on” detection of Uric acid, glucose and alcohol in sweat. Biosens. Bioelectron. 192, 113530 (2021).
Lee, H. et al. A graphene-based electrochemical device with thermoresponsive microneedles for diabetes monitoring and therapy. Nat. Nanotechnol. 11, 566–572 (2016).
Zhao, J. et al. A wearable nutrition tracker. Adv. Mater 33, 2006444 (2021).
Tai, L. C. et al. Wearable sweat band for noninvasive levodopa monitoring. Nano Lett. 19, 6346–6351 (2019).
Bariya, M. et al. Roll-to-roll gravure printed electrochemical sensors for wearable and medical devices. ACS Nano 12, 6978–6987 (2018).
He, X. et al. Integrated smart janus textile bands for self-pum** sweat sampling and analysis. ACS Sens. 5, 1548–1554 (2020).
Sun, M. et al. A flexible microfluidic chip-based universal fully integrated nanoelectronic system with point-of-care raw sweat, tears, or saliva glucose monitoring for potential noninvasive glucose management. Anal. Chem. 94, 1890–1900 (2022).
Zong, C. et al. Surface-enhanced Raman spectroscopy for bioanalysis: reliability and challenges. Chem. Rev. 118, 4946–4980 (2018).
Wang, Z., Zong, S., Wu, L., Zhu, D. & Cui, Y. SERS-activated platforms for immunoassay: probes, encoding methods, and applications. Chem. Rev. 117, 7910–7963 (2017).
Liu, Y., Kim, M., Cho, S. H. & Jung, Y. S. Vertically aligned nanostructures for a reliable and ultrasensitive SERS-active platform: fabrication and engineering strategies. Nano Today 37, 101063 (2021).
Xu, K., Zhou, R., Takei, K. & Hong, M. Toward flexible surface‐enhanced Raman scattering (SERS) sensors for point‐of‐care diagnostics. Adv. Sci. 6, 1900925 (2019).
Jeong, J. W. et al. 3D cross-point plasmonic nanoarchitectures containing dense and regular hot spots for surface-enhanced Raman spectroscopy analysis. Adv. Mater. 28, 8695–8704 (2016).
Andreou, C., Hoonejani, M. R., Barmi, M. R., Moskovits, M. & Meinhart, C. D. Rapid detection of drugs of abuse in saliva using surface enhanced Raman spectroscopy and microfluidics. ACS Nano 7, 7157–7164 (2013).
Wang, Y. et al. Wearable plasmonic-metasurface sensor for noninvasive and universal molecular fingerprint detection on biointerfaces. Sci. Adv. 7, eabe4553 (2021).
Kim, W. et al. Paper-based surface-enhanced Raman spectroscopy for diagnosing prenatal diseases in women. ACS Nano 12, 7100–7108 (2018).
Park, J. E. et al. Plasmonic microneedle arrays for in situ sensing with surface-enhanced Raman spectroscopy (SERS). Nano Lett. 19, 6862–6868 (2019).
Koh, E. H. et al. A wearable surface-enhanced Raman scattering sensor for label-free molecular detection. ACS Appl. Mater. Interfaces 13, 3024–3032 (2021).
Chung, M. et al. Fabrication of a wearable flexible sweat pH sensor based on SERS-active Au/TPU electrospun nanofibers. ACS Appl. Mater. Interfaces 13, 51504–51518 (2021).
Sevim, S. et al. SERS barcode libraries: a microfluidic approach. Adv. Sci. 7, 1903172 (2020).
Mogera, U. et al. Wearable plasmonic paper-based microfluidics for continuous sweat analysis. Sci. Adv. 8, eabn1736 (2022).
Ding, S. Y., You, E. M., Tian, Z. Q. & Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 46, 4042–4076 (2017).
Sikirzhytski, V., Sikirzhytskaya, A. & Lednev, I. K. Multidimensional Raman spectroscopic signature of sweat and its potential application to forensic body fluid identification. Anal. Chim. Acta 718, 78–83 (2012).
Yang, Y. et al. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 38, 217–224 (2020).
Brooks, G. A. The science and translation of lactate shuttle theory. Cell Metab. 27, 757–785 (2018).
Gao, W. et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature 529, 509–514 (2016).
Yu, Y. et al. Biofuel-powered soft electronic skin with multiplexed and wireless sensing for human-machine interfaces. Sci. Robot. 5, eaaz7946 (2020).
Jiang, T. et al. Generation and phenotypic analysis of mice lacking all urea transporters. Kidney Int. 91, 338–351 (2017).
Dai, Z., Li, Y., Duan, G., Jia, L. & Cai, W. Phase diagram, design of monolayer binary colloidal crystals, and their fabrication based on ethanol-assisted self-assembly at the air/water interface. ACS Nano 6, 6706–6716 (2012).
Johnson, P. B. & Christy, R. W. Optical constants of the noble metals. Phys. Rev. B 6, 4370–4379 (1972).
Rott, N. Note on the history of the Reynolds number. Annu. Rev. Fluid Mech. 22, 1–12 (1990).
Wilke, C. & Chang, P. Correlation of diffusion coefficients in dilute solutions. AICHE J. 1, 264–270 (1955).
Acknowledgements
We acknowledge funding from Joint Fund of the Ministry of Education for Equipment Pre-research (8091B022142), Shenzhen Stability Support Plan (20200806163622001), Shenzhen Overseas Talent Program, and Shenzhen Key Laboratory for Nano-Biosensing Technology (ZDSYS20210112161400001). We also appreciate the measurement equipment of portable Raman analyzer (including hardware and software support) provided by Optosky Photonics Inc, China.
Author information
Authors and Affiliations
Contributions
H.X., F.C., X.T., and Z.X. conceived and designed the experiments. H.X. and L.Y. prepared materials, performed the experiments, and analyzed experimental data. H.X. wrote the manuscript. X.T. revised the manuscript. X.T. and Z.X. supervised all the aspects of this work and provided financial support. All authors discussed the results and contributed to the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, X., Fan, C., Luo, Y. et al. Flexible microfluidic nanoplasmonic sensors for refreshable and portable recognition of sweat biochemical fingerprint. npj Flex Electron 6, 60 (2022). https://doi.org/10.1038/s41528-022-00192-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41528-022-00192-6
- Springer Nature Limited
This article is cited by
-
Epidermal wearable optical sensors for sweat monitoring
Communications Materials (2024)
-
Wearable flexible microfluidic sensing technologies
Nature Reviews Bioengineering (2023)
-
Large-scale fully printed “Lego Bricks” type wearable sweat sensor for physical activity monitoring
npj Flexible Electronics (2023)