Abstract
Crop wild relatives offer natural variations of disease resistance for crop improvement. Here, we report the isolation of broad-spectrum powdery mildew resistance gene Pm36, originated from wild emmer wheat, that encodes a tandem kinase with a transmembrane domain (WTK7-TM) through the combination of map-based cloning, PacBio SMRT long-read genome sequencing, mutagenesis, and transformation. Mutagenesis assay reveals that the two kinase domains and the transmembrane domain of WTK7-TM are critical for the powdery mildew resistance function. Consistently, in vitro phosphorylation assay shows that two kinase domains are indispensable for the kinase activity of WTK7-TM. Haplotype analysis uncovers that Pm36 is an orphan gene only present in a few wild emmer wheat, indicating its single ancient origin and potential contribution to the current wheat gene pool. Overall, our findings not only provide a powdery mildew resistance gene with great potential in wheat breeding but also sheds light into the mechanism underlying broad-spectrum resistance.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) is the world’s most widely grown food crop, providing 20% of the total daily calories and protein in human nutrition1. Pathogens and pests have been imposing big challenges on wheat production, therefore threatening global food security. Wheat powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is one of the most devastating diseases causing dramatic reductions in both grain yield and quality over the past decades across the world2. Improving crop resistance to pathogens through breeding is an effective and environmentally sound strategy for managing disease and minimizing these losses. However, the emergence of new virulent pathogen populations always makes resistant wheat varieties become susceptible3. Therefore, resistance breeding is always like an ‘arm race’ and looking for broad-spectrum and durable disease resistance genes is still needed.
To date, eighteen powdery mildew resistance (Pm) genes, Pm1a4, Pm25, Pm36, Pm47, Pm5e30. We previously mapped a powdery mildew resistance gene Ml3D232, derived from WEW in the hexaploid wheat introgression line 3D232, on the overlap** genetic interval with Pm3631. We then deduced that they are likely the same gene or alleles by comparative genetic linkage maps32. However, the molecular relationship of the two genes is still unclear.
In this study, we report map-based cloning of Ml3D232/Pm36, originated from WEW and confers broad-spectrum resistance to Bgt in both durum and common wheat. Pm36 encodes a unique tandem kinase with a transmembrane domain in the C-terminus (WTK7-TM). We demonstrate that the two kinase domains (Kin I and Kin II) and the transmembrane domain of WTK7-TM are critical for its powdery mildew resistance function. We also show that both Kin I and Kin II are indispensable for the kinase activity of WTK7-TM. Pm36 is only present in a few WEW populations with a single ancient origin and can potentially contribute to modern wheat improvement.
Results
Fine map** of Ml3D232
WEW-hexaploid wheat introgression line 3D232 exhibits a broad-spectrum resistance against an extensive collection of tested 104 genetically distinct Bgt isolates at the seedling stage, including the most prevailing virulent isolates E09, E20, and E21 from China (Supplementary Fig. 1 and Supplementary Data 1). 3D232 is also highly resistant to Bgt isolate E09 at the adult plant stage under the field condition (Supplementary Fig. 2). The powdery mildew resistance gene Ml3D232 in 3D232 was previously identified and mapped on chromosome arm 5BL31. Using comparative genetic linkage map**, we found the map** interval of Ml3D232 overlapped with Pm36 derived from WEW, indicating that they are likely the same gene or allelic32.
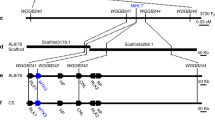
To find map** Ml3D232 gene, two Ml3D232-flanking simple sequence repeats (SSR) markers XBD37670 and XBD37760 on chromosome 5BL32 were selected to screen 15,893 individual F2 plants derived from the cross between a highly susceptible common wheat line Xuezao and the resistant introgression line 3D232 (Fig. 1a and Supplementary Data 2). Finally, Ml3D232 was narrowed down to a genetic interval of 0.021-cM flanked by the closest derived cleaved amplified polymorphic sequence (dCAPS) marker WGGBM2 and SSR marker WGGBM7 (Fig. 1b and Supplementary Data 2), which corresponds to 273.8 kb genomic region of wild emmer wheat Zavitan (WEW_v1.0)33 and contains four predicted high-confidence genes, a cytochrome B561 and domon domain protein (CYB561-Domon), two serpin proteins (Serpin 1 and Serpin 2), and a transmembrane protein (TM9SF4) (Fig. 1c and Supplementary Data 2). qRT-PCR analyses showed that CYB561-Domon and TM9SF4 were constitutively expressed in leaves of 3D232 and Xuezao seedlings, regardless of inoculation with Bgt isolate E09, whereas no expression of the two serpin genes was detected in either 3D232 or Xuezao (Supplementary Fig. 3a, b). Since differential expressions and coding sequence, polymorphisms were observed for CYB561-Domon (Supplementary Figs. 3 and 4) and TM9SF4 (Supplementary Figs. 3 and 5) genes between 3D232 and Xuezao, two overexpression constructs, ProUbi: CYB561-Domon and ProUbi: TM9SF4, consist of the coding sequences (CDS) of the two genes from 3D232 and driven by the maize (Zea mays L.) Ubiquitin promoters (Supplementary Fig. 6a, b), were separately delivered into highly susceptible wheat cultivar Fielder by Agrobacterium-mediated transformation. Unfortunately, all plants positive for ProUbi: CYB561-Domon and ProUbi: TM9SF4 constructs in the independent T1 transgenic families were highly susceptible to the Bgt isolate E09 (Supplementary Fig. 6c–e), suggesting CYB561-Domon and TM9SF4 are not the causal gene of Ml3D232.
a Infection types of line 3D232 and Xuezao to Bgt isolate E09. Plants of 3D232 and Xuezao were inoculated with Bgt isolate E09 at the two-leaf stage. Plant leaves were detached and photographed at 10 day post-inoculation (dpi). Scale bar, 3 mm (1). Fungal structures of Bgt isolate E09 at 72 h post inoculation (hpi) were stained by Coomassie Brilliant Blue. Scale bar, 100 μm (2). DAB-Coomassie blue staining of infected leaves with Bgt isolate E09 from 3D232 and Xuezao at 48 hpi. Brown staining shows the accumulation of H2O2. Scale bar, 100 μm (3). Line 3D232 is immune to Bgt isolate E09, while Xuezao is highly susceptible to Bgt isolate E09. Three independent experiments were performed. b Genetic map of the region carrying Ml3D232 on the long arm of Chinese Spring chromosome 5B. c Physical map covering the Ml3D232 region on Zavitan reference genome (WEW_v1.0). d Physical map covering the Ml3D232 region on tetraploid WEW-durum introgression line 5BIL-29 genome. e Protein prediction of wheat tandem kinase with single transmembrane (WTK7-TM). The kinase I (Kin I), kinase II (Kin II), and transmembrane (TM) domains of WTK7-TM are highlighted in orange, yellow, and cyan circles respectively. Source data are provided as a Source Data file.
PacBio SMRT long-read sequencing of tetraploid WEW-durum introgression line 5BIL-29
In order to isolate the causal gene underpinning locus Ml3D232/Pm36, we selected the tetraploid WEW-durum introgression line 5BIL-29 carrying Pm3630 for sequencing through the PacBio SMRT long-read sequencing approach. PacBio CCS (HiFi) reads corresponding to approximately 19.4-fold coverage of tetraploid wheat genome were generated (Supplementary Data 3). In addition, about 117.3-fold Hi-C reads were added to the assembly. After sequence assembling, 2207 contigs were obtained, with a contig N50 of 20.8 Mb and N90 of 4.5 Mb, the longest contig length of 168.3 Mb, and the total length of the genome assembly of 10.5 Gb, which covered the whole genome of WEW Zavitan33 and durum wheat Svevo34 (Supplementary Data 3).
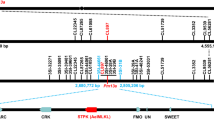
A 7.1 Mb contig ptg000422 spanning the entire Pm36 physical map** interval (1.17 Mb) was captured by aligning the flanking and co-segregating markers WGGBM2 to WGGBM7 around the Pm36 locus (Fig. 1d). There were eleven annotated genes in the 1.17 Mb physical interval of Ml3D232/Pm36 between the closest flanking markers WGGBM2 and WGGBM7: CYB561-Domon, three F-box proteins (F-box 1-3), five Serpin proteins (Serpin 1-5), a tandem kinase protein with a single transmembrane domain in the C-terminus (WTK7-TM), and TM9SF4 (Fig. 1d). Micro-collinearity analysis revealed highly conserved protein-coding genes in the Pm36 genomic region among multiple wheat reference genomes33,34,35,36,37, except for the WEW-durum introgression line 5BIL-29 with significant genomic structural variation (Supplementary Figs. 7 and 8). Compared to Zavitan, and all other available reference genomes, introgression line 5BIL-29 contains a ~ 900 kb insertion in the Pm36 physical region harboring two segmental duplications containing 7 predicted genes (Fig. 1c, d; Supplementary Figs. 7, 8, and Supplementary Data 4). An about 70 kb fragment containing one F-box and two Serpin genes was likely duplicated twice. The first duplication was nearly intact with an additional 22 kb insertion resulting in 92 kb sequence. The second duplication contains the F-box, only one Serpin, the traces of 70 kb transposable elements, and a 300 kb sequence harboring the WTK7-TM (Supplementary Fig. 9). RNA-seq analysis showed that only CYB561-Domon, TM9SF4, and WTK7-TM were expressed among the eleven annotated genes in the 1.17 Mb genomic interval in the leaves of 5BIL-29 seedlings after inoculation with Bgt isolate E09 (Supplementary Data 5). Given that CYB561-Domon and TM9SF4 have been previously excluded by transgenic assay (Supplementary Fig. 6c), WTK7-TM was most likely the candidate gene for Ml3D232/Pm36.
Cloning of WTK7-TM
We then cloned the 10,741 bp genomic DNA fragment of WTK7-TM including 3277 bp coding region, 3311 bp promoter region, and 4153 bp terminator sequence from lines 5BIL-29 and 3D232, and found no sequence variation in WTK7-TM between lines 5BIL-29 and 3D232. The full-length cDNAs of WTK7-TM were also obtained from line 3D232 by the 5′- and 3′-rapid amplification of complementary DNA (cDNA) ends (RACE). Nine transcription isoforms of WTK7-TM, named as TV1-TV9, were identified, with isoform TV1 being the most common among them (Supplementary Fig. 10). The WTK7-TM gene has 12 exons, which encodes a 667-residue tandem kinase protein including Kinase I (Kin I) and Kinase II (Kin II) domains, and a predicted single transmembrane domain in the C-terminus (Fig. 1e and Supplementary Fig. 11). Moreover, we found that WTK7-TM is constitutively expressed in the root, stem, and leaf tissues without Bgt infection (Supplementary Fig. 12a), but could be slightly upregulated after Bgt isolate E09 infection in lines 3D232 and 5BIL-29 (Supplementary Fig. 12b), prioritizing it for the further validation as a candidate gene of Ml3D232/Pm36.
Validation of WTK7-TM by VIGS, EMS mutants, and transgenic assay
To examine whether WTK7-TM is responsible for the powdery mildew resistance of Ml3D232/Pm36 in 3D232 and 5BIL-29, we performed virus-induced gene silencing (VIGS) experiment38 with two constructs targeting the regions of Kin I and Kin II domains of WTK7-TM, respectively (Supplementary Fig. 13). The transcript levels of WTK7-TM in lines 3D232 and 5BIL-29 were substantially knocked down in both BSMV:WTK7-TMKin I and BSMV:WTK7-TMKin II infected plants compared to BSMV:00 infected plants (Fig. 2a and Supplementary Fig. 13). As a result, both lines 3D232 and 5BIL-29 lost their resistance to Bgt isolate E09 (Fig. 2b). The results indicated that WTK7-TM is necessary for resistance to Bgt in lines 3D232 and 5BIL-29.
a Relative expression levels of WTK7-TM in lines 3D232 and 5BIL-29 treated with BSMV:00 (control), BSMV:WTK7-TMKin I and BSMV:WTK7-TMKin II, respectively, were examined by qRT-PCR. Results represent the means ± SD from three biological replicates (three leaves used per biological replicate) and three technical replicates for each leave (N = 9, unpaired two-tailed t-test). “****” indicates a statistically significant difference (P < 0.0001). b Plants were inoculated with Bgt isolate E09 and representative leaves were photographed at 14 dpi. VIGS1 and VIGS2 showed the two target positions of WTK7-TM in BSMV-VIGS experiments. Three independent experiments were performed. c Line 3D232 and 17 susceptible mutants on WTK7-TM inoculated with Bgt isolate E09 and representative leaves were photographed at 14 dpi. Three independent experiments were performed. d Susceptible EMS mutants carrying single nonsense or missense mutations in the WTK7-TM gene. Black straight lines indicate introns, and rectangles indicate coding exons. Domains Kin I and Kin II, and the transmembrane region of WTK7-TM are highlighted in orange, yellow, and cyan rectangles, respectively. Short vertical lines indicate the mutated positions. The DNA sequence (D.) changes, and their predicted effects on the translated protein (P.) are indicated below the mutants. Mutation names in red, purple, and black indicate nonsense, splice site, and missense mutations, respectively. P-values and source data are provided as a Source Data file.
To further validate the function of WTK7-TM, we used ethyl methane sulfonate (EMS) to mutagenize line 3D232 and identified 23 M3 mutants highly susceptible to Bgt isolate E09 out of 3360 M3 families (Fig. 2c and Supplementary Data 6). Further characterization with PCR and Sanger sequencing of the WTK7-TM gene including the promoter sequence, coding region, and 3’ downstream sequence indicated that 17 mutants carried mutations within the CDS of WTK7-TM, including five premature stop codons, one frameshift alternative splicing variant, and 11 missense mutations (Fig. 2d and Supplementary Data 6). No sequence variations were detected at the WTK7-TM in the other 6 mutants, suggesting other factors may be involved in the regulation of WTK7-TM resistance. Seven mutants had mutations in the conserved Kin I domain and 8 mutants showed mutations in the Kin II domain, indicating that both Kin I and Kin II domains are essential for the function of WTK7-TM in conferring resistance to powdery mildew (Fig. 2d). Surprisingly, the loss of function mutant M16 carried a nonsense mutation in the transmembrane domain, generating a premature WTK protein without the transmembrane domain (Fig. 2c, d; Supplementary Fig. 14 and Supplementary Data 6). A point mutation was detected in mutant M17 in the splice acceptor site of intron 11, resulting in the frameshift alternative splicing manner with an extra 9 amino acids after the transmembrane domain (Fig. 2c, d; Supplementary Fig. 14 and Supplementary Data 6). This result demonstrated the importance of the transmembrane domain for the function of WTK7-TM. These independent mutations demonstrate that Kin I, Kin II, and TM domains of WTK7-TM are required for resistance against Bgt.
To further determine whether WTK7-TM is Ml3D232/Pm36, we cloned a 10,741 bp genomic fragment of WTK7-TM from 3D232, comprising the complete gene coding region and its regulatory elements, into the binary vector pCAMBIA-1300 (Fig. 3a) and transformed the susceptible wheat cultivar Fielder by Agrobacterium-mediated transformation. We obtained five independent T0 transgene-positive plants by PCR analysis, which were self-pollinated and advanced to the T1 generation. Evaluation of T1 progeny of transgenic lines showed that individuals carrying the WTK7-TM transgene were resistant to Bgt isolate E09 (Fig. 3b, c). Taken together, these results show that WTK7-TM is the Ml3D232/Pm36 gene.
a Structure of ProWTK7-TM:WTK7-TM used for transgenic assay. The ProWTK7-TM:WTK7-TM construct contains the 3277 bp full-length WTK7-TM coding region, as well as 3311 bp upstream of the start codon and 4153 bp downstream of the stop codon. LB, left border; RB, right border. b The resistance performance of ProWTK7-TM:WTK7-TM–transformed transgenic plants. Transgenic T1 wheat (about 15 plants per family) was challenged with Bgt isolate E09 at the two-leaf stage. Representative leaves were photographed at 12 dpi. PCR results as positive (+) or negative (-) for DNA amplification (upper) and RNA expression (lower) of the WTK7-TM gene. Three independent experiments were performed. c The relative expression levels of WTK7-TM in transgenic wheat. Leaves of seedling plants at the two-leaf stage were collected. TaActin was used as an endogenous control. Expression values are means ± SD from three biological replicates (three leaves used per biological replicate) three biological replicates (three leaves used per biological replicate) and three technical replicates for each leave (N = 9). There was no expression in non-transgenic plants. Error bars are standard errors of the mean. Source data are provided as a Source Data file.
Subcellular localization, structure, and catalytic activity of WTK7-TM
Recent studies revealed that TKPs are key regulators of crop pathogen resistance, especially in wheat and barley23. However, all reported TKPs did not contain any membrane-targeting motif, except for WTK7-TM, suggesting a possible different resistance mechanism for this protein (Supplementary Fig. 15). Subcellular localization displayed WTK7-TM protein is localized in the plasma membrane in N. benthamiana (Supplementary Fig. 16). Phylogenetic analysis showed that both kinase domains of WTK7-TM are located in the same clade with Rpg1, WTK4, and WTK6-vWA that originate through kinase domain duplication, suggesting a possible duplication origin of WTK7-TM (Fig. 4). Based on Hidden Markov Model, both the two kinase domains of WTK7-TM belong to LRR_8B (cysteine-rich kinases) subfamily, like as Rpg124. Further analysis by aligning WTK7-TM kinase domains with well-studied kinases indicated that Kin I and the Kin II domains of WTK7-TM are serine/threonine (S/T) non-arginine aspartate (non-RD) kinase with potentially catalytic activity, while two kinase domains are more likely pseudokinase due to the loss of some conserved residues (Supplementary Fig. 17). The three-dimensional (3D) model of WTK7-TM protein predicted by AlphaFold v2.039 revealed the Kin I and Kin II domains, and a single transmembrane domain in the C-terminus (Fig. 5a). Possessing two tandem kinase domains is the most distinct feature of TKPs, however, little is known on how these TKPs perform their kinase activities23. We, therefore, explored the WTK7-TM in vitro phosphorylation activity. The results showed that the full-length WTK7-TM (1–667 aa), Kin I (1–320 aa) or Kin II (321–646 aa) domain alone doesn’t exhibit any auto-phosphorylation or catalytic activity, while the truncated version WTK7-TMKin I-Kin II (1–646 aa) composing of Kin I + Kin II without the transmembrane domain does demonstrate such activities (Fig. 5b, c and Supplementary Fig. 18).
a WTK7-TM protein structure was predicted on AlphaFold v2.0. Domains Kin I and Kin II, and the transmembrane region of WTK7-TM are highlighted in orange, yellow, and cyan, respectively. Mutations on domains Kin I and Kin II of WTK7-TM are indicated by red spheres. b Schematic diagram of the full-length protein WTK7-TM (1–667 aa), Kin I (1–320 aa), Kin II (321–646 aa) and WTK7-TMKin I-Kin II consisting of Kin I + Kin II without transmembrane of WTK7-TM (1–646 aa). c Kinase activity analysis of truncated WTK7-TM in vitro. Neither Kin I nor Kin II domains have kinase activity while WTK7-TMKin I-Kin II (1–646 aa) has auto-phosphorylation and can phosphorylate myeline basic protein. Autoradiograph and coomassie brilliant blue staining are shown in the top and bottom panels, respectively. GST-GFP was used as a substrate for negative control. Three independent experiments were performed. Source data are provided as a Source Data file.
Geographic distribution of WTK7-TM
To evaluate the distribution of WTK7-TM in wild and cultivated wheat, we developed a functional marker WTK7-TM-FM for specific detection of WTK7-TM and screened wheat germplasm collected from geographically diverse sites in the world, including WEW (442 accessions), cultivated emmer (125 accessions), durum (230 accessions), and hexaploid wheat (1292 accessions) (Supplementary Data 7). WTK7-TM was detected only in 38 WEW accessions, which were resistant to Bgt isolates E09 and E20 (Supplementary Fig. 19 and Supplementary Data 8) and shared an identical genomic sequence and CDS of WTK7-TM. All of these WEW accessions originated from the southern (Israel, Lebanon, Jordan, and Syria) WEW populations and represent only 1.82% of the tested wheat germplasms (Supplementary Fig. 20). The absence of WTK7-TM from the other WEW populations, as well as domesticated emmer, durum, and common wheat suggests that WTK7-TM is likely an orphan gene with single ancient origin and will have a profound contribution to the current wheat gene pool.
Discussion
Most characterized plant resistance proteins belong to the race-specific nucleotide-binding domain and leucine-rich repeat-containing (NLR) family that recognizes pathogen race-specific effectors40. Recently, a class of intracellular resistance proteins with tandem kinase domain architecture has emerged23 and provides resistance against not only biotrophic fungal pathogens, Puccinia graminis f. sp. tritici (Pgt)24,26,27, P. striiformis f. sp. tritici (Pst)25, P. triticina f. sp. tritici (Pt)28 and B. graminis f. sp. tritici (Bgt)15,19,22, but also hemibiotrophic pathogen, Pyricularia oryzae (Po)29. Here, we isolated a broad-spectrum powdery mildew resistance gene Pm36 (WTK7-TM) originated from wild emmer wheat by combining map-based cloning and PacBio SMRT long-read genome sequencing approach. WTK7-TM encodes a tandem kinase protein with a single transmembrane domain in its C-terminus. To date, all functionally validated tandem kinase proteins in plants did not contain any membrane-targeting motif23, except for WTK7-TM, suggesting a possible different resistance mechanism for this unique protein.
In plants, membrane-related proteins play essential roles in response to pathogens. Receptor-like proteins (RLPs) or receptor-like kinases (RLKs) containing a transmembrane (TM) domain and extracellular LRR domains, either a short domain or a kinase domain, respectively, can recognize microbe - or plant-derived immunogenic molecular patterns from outside of the plant cell, resulting in the activation of immune responses41. Arabidopsis thaliana broad-spectrum mildew resistance gene Rpw8 is specifically targeted to the extrahaustorial membrane where it promotes H2O2 accumulation and triggers a hypersensitive response to constrain the haustorium47. Although multiple wheat genome sequences and abundant variations are already available33,34,35,36,37, cloning of agronomically important genes, including disease resistance genes, remains a challenging task at a particular locus. Physical map** and sequence comparison indicated an approximate 136 kb fragment deletion was present in the corresponding Pm41 genomic region in Chinese Spring chromosome 3BL17. Maize ZmWUS1 locus exists a large tandem segmental duplication event that created a unique 119 bp insertion in the proximal promoter region of the duplicate ZmWUS1-B copy48. In our study, we sequenced a tetraploid WEW-durum wheat introgression line 5BIL-29 carrying Pm36 using PacBio SMRT long-read sequencing approach. After sequence assembly, we found that Pm36 locus underwent a ~ 900 kb insertion harboring an additional 7 annotated genes including WTK7-TM with two segmental duplications compared to Zavitan and other available wheat reference genomes. The finding also demonstrates structural variations are major obstacles to gene identification in complex genomes, such as wheat and wild relatives, based on the limited availability of reference and pan-genome sequences. Recently, long-read sequencing technologies have also been used to clone wheat yellow rust resistance gene Yr27 49 and powdery mildew resistance gene Pm6921, indicating a powerful tactic for facilitating disease resistance gene cloning in wheat.
The crop wild relatives serve as an important untapped reservoir of genetic diversity that can be used to improve modern breeding. The process of wheat domestication and hexaploidization has significantly reduced genetic diversity relative to the wild ancestor across the genome50. WTK7-TM was found only in the southern WEW populations and seems to be absent in other WEW populations, as well as domesticated emmer, durum, and common wheat, indicating a relatively ancient single origin. The geographical distribution of WTK7-TM is similar with Pm4117, Yr1525, and Yr3651 appearing in the Mount Carmel to Anti-Lebanon mountain ridge, manifesting host–parasite coevolution in WEW natural habitats. Although more than a hundred Pm genes have been reported, only a few can provide resistance to most or all Bgt isolates, such as Pm12 from Aegilops speltoides10, Pm2113,31. A single dominant gene Ml3D232 conferring powdery mildew resistance has been mapped to the chromosome arm 5BL (bin 5BL 0.59–0.76) using a molecular and deletion map** strategy31. A backcross inbred line 5BIL-29 carrying powdery mildew resistance gene Pm36 was previously developed by crossing wild emmer wheat accession MG29896 with durum wheat cv. Latino30 and kindly provided by Dr. Antonio Blanco, Department of Soil, Plant and Food Sciences (DiSSPA), Genetics and Plant Breeding Section, University of Bari Aldo Moro, Italy. Wheat collections for geographical distribution analysis of Ml3D232/Pm36 included 442 wild emmer wheat (T. turgidum ssp. dicoccoides), 125 cultivated emmer wheat (T. turgidum subsp. dicoccum), 230 durum (T. turgidum subsp. durum), 262 entries of Chinese wheat mini-core collection (MCC)53, 636 and 394 common wheat accessions from China and the other 34 countries, respectively55. Genetic markers linked to Ml3D232/Pm36 including SSR and dCAPs markers (Supplementary Data 2) were developed using the Chinese Spring (RefSeq v1.0)35 and the WEW accession Zavitan reference genome (WEW_v1.0)33 sequences for constructing a high-resolution genetic linkage map. The F2 population from the Xuezao × 3D232 cross was first genotyped with SSR markers XBD37670 and XBD37760, which flank Ml3D23232, and individuals with recombination events between these markers were then genotyped with all the other markers (Supplementary Data 2) and subjected to powdery mildew assay for the F3 progenies. The linkage between genetic markers and Ml3D232 was established using JoinMap4.0 (http://www.kyazma.nl/index.php/mc.Join-Map/sc.General/) with a LOD threshold of 3.0. Genetic map distances were calculated by means of the Kosambi map** function56.
PacBio HiFi and Hi-C sequencing
High molecular weight DNA was isolated from 20-day-old fresh leaf tissue of 5BIL-29 carrying Pm36 following the large-scale nucleus extraction protocols57. DNA purity was assessed based on the ratio of absorbance at 260 nm and 280 nm (A260/A280) using a NanoDrop NP-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). DNA yield was assessed using the Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Waltham, USA), and DNA size was validated by pulsed-field gel electrophoresis (PFGE) of Pippin pulse. DNA was sheared to the appropriate size range (15–20 kb) using a Covaris g-TUBE for the construction of PacBio HiFi sequencing libraries58, followed by bead purification with PB Beads (PacBio). HiFi sequencing libraries were prepared using SMRTbell Express Template Prep Kit 2.0 and followed by immediate treatment with the Enzyme Clean Up Kit. The libraries were further size-selected electrophoretically using BluePippin Systems from SAGE Science. Sequencing was performed on the Pacific Biosciences Sequel II system using HiFi sequencing protocols. Libraries were sequenced on six PacBio SMART cells (Supplementary Data 3).
For chromosome conformation capture (Hi-C) sequencing, isolated DNA from 20-day-old 5BIL-29 fresh leaf tissue was cross-linked and fixed. The fixed samples were sent to Wuhan Grandomics Biosciences Co., Ltd, China, for Hi-C library construction and sequencing. The quality of the libraries was assessed with Agilent 2100 Bioanalyzer (Agilent), and subjected to 2 × 150 bp paired-end high-throughput sequencing by Illumina NovaSeq/MGI. Finally, 1220.8 Gb of effective data were obtained (Supplementary Data 3).
Genome assembly and gene annotation
The cumulative length of subreads from each of the six PacBio SMART cells ranged from 455.6 to 530.3 Gb, resulting in a combined length of HiFi reads ranging from 29.61 to 35.26 Gb, total 202.14 Gb. The N50 for HiFi reads ranged from 15.9 to 23.2 kb, with the max subreads length 65 kb. All the HiFi and Hi-C reads were assembled using the default parameters of Hifiasm 0.16.1-r37559, and the gfatools (https://github.com/lh3/gfatools) was used to convert sequence graphs in the GFA to FASTA format. We captured a 7.1 Mb contig ptg000422l using flanking and co-segregating markers (WGGBM2 to WGGBM7) of Ml3D232, which can entirely cover the Ml3D232/Pm36 physical map** interval (1.17 Mb). This 1.17 Mb genomic sequence was annotated using Softberry FGENESH60 and NCBI BLASTP against the nr Database to identify homologous proteins. Repetitive sequences and transposable elements (TEs) were identified through BLASTN against the Triticeae repetitive elements (TREP) (https://trep-db.uzh.ch/). The long terminal repeat sequences of LTR retrotransposable elements were delineated using Dotter analysis and manually checked. The evolutionary distance of the two LTR sequences was estimated by MEGA11 (https://www.megasoftware.net/) using a substitution rate of 1.3 × 10−8 mutations per site per year.
RNA-seq, quantitative real-time PCR, and RT-PCR
Total RNA samples were extracted from two-leaf stage seedling leaves of 3D232 and 5BIL-29 at 0, 6, 12, 24, 36, 48, and 72 hpi with Bgt isolate E09 using the TRIzol reagent (Tiangen, Bei**g, China). The first-strand cDNA from total RNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Kyoto, Japan), and for quantitative real-time PCR (qRT-PCR) was performed with SYBR green kit (TaKaRa, Kyoto, Japan) in a Roche 480 light cycler (Roche, Colorado Springs, CO, USA). TaActin was used as the internal control, and the 2−ΔΔCt method was used to calculate relative gene expression61. To investigate whether WTK7-TM is expressed in different wheat tissues, we conducted RT-PCR analysis in the leaf, stem, and root of 3D232 at the two-leaf stage. TaActin was used as the endogenous control. Three biological replicates (three leaves or roots or stems used per biological replicate) from three independent experiments and three technical replicates for each sample were performed.
For RNA-seq, the RNA samples from two-leaf stage seedling leaves of 3D232 or 5BIL-29 inoculated with Bgt isolate E09 at 24hpi were selected to construct cDNA libraries and subjected to high-throughput paired-end sequencing by Illumina HiSeq2000 at Bei**g Novogene Bioinformatics Technology Co. Ltd, Bei**g, China. Clean reads were aligned to the reference genome of Chinese Spring or contig ptg000422l by Hisat262, and SNPs were called by GATK haplotypeCaller63 following the GATK best practice for RNA-Seq.
3′- and 5′-RACE
Total RNA was isolated from two-leaf stage seedling leaves of 3D232 inoculated with Bgt isolate E09 at 24 hpi, using the TRIzol reagent (Tiangen, Bei**g, China). Reverse transcription was performed using a PrimeScript RT reagent Kit with a gDNA Eraser (TaKaRa, Kyoto, Japan). We perform a rapid amplification of cDNA ends (RACE) to determine the transcriptional start (5′-RACE) and end (3′-RACE) of the WTK7-TM gene. The 3′- and 5′-UTR sequences of WTK7-TM were identified using the SMARTer RACE cDNA Amplification Kit (cat. no. 634923; Clontech, Mountain View, CA, USA). One hundred colonies were sequenced using the Sanger sequencing.
Virus-induced gene silencing
We created gene-silencing constructs by cloning fragments of WTK7-TM into the BSMV RNA γ-derived binary vector pCa-γbLIC38. Two fragments of WTK7-TM, one from the Kin I domain and the other from the Kin II domain were amplified from recombinant vector pEASY:WTK7-TM TV1 carrying TV1 transcription isoform of WTK7-TM and inserted separately into pCa-γbLIC to generate independent recombinant constructs, BSMV:WTK7-TMKin I, and BSMV:WTK7-TMKin II. Firstly, equal amounts of Agrobacterium tumefaciens strain GV3101 harboring pCaBS-α, pCaBS-β and pCa-γbLIC (or BSMV:WTK7-TMKin I, and BSMV:WTK7-TMKin II) were mixed and infiltrated into N. benthamiana leaves at four to eight-leaf stage with a 1-mL needleless syringe. At 10 dpi, the infiltrated N. benthamiana leaves were ground in 20 mM Na-phosphate buffer (pH 7.2) containing 1% celite38, and the sap was mechanically inoculated onto 3D232 and 5BIL-29 leaves at the two-leaf stage. Three weeks after inoculation with the N. benthamiana leaf sap, wheat develo** visible BSMV symptoms (stripe mosaic on the 3th leaf with a mild mosaic on the 4th leaf) was challenged with Bgt isolate E09. Powdery mildew reactions of each plant were rated on a 0–4 IT scale15. The expression levels of WTK7-TM were also determined by qRT-PCR.
Ethyl methane sulfonate mutagenesis and screening
Ethyl methane sulfonate (EMS) mutagenesis of 3D232 was performed with a concentration of 0.6% EMS (M0880, Sigma-Aldrich, Shanghai, China). Seeds were soaked in water for 12 h, incubated for 16 h with shaking in EMS solution and washed with running water for 4 h at room temperatures64. Mutagenized seeds were grown in the field at Gaoyi Experimental Station, Shijiazhuang, Hebei province, and 3360 M1 plants were harvested. M2 families (15–25 seeds per family) were screened for their response to Bgt isolate E09 under glasshouse conditions. Susceptible M2 plants were transplanted and grown in pots and the M3 progenies were challenged again with Bgt isolate E09 to confirm their susceptibility, as well as for authenticity of genotype using an array of SSR markers (Supplementary Data 2). Finally, we obtained 23 independently susceptible M3 mutants of 3D232. The full-length genomic sequence of the WTK7-TM including the promoter, coding region, and terminator sequence from each of the 23 mutants was obtained using the primers, WTK7-60, WTK7-61, and WTK7-62 listed in Supplementary Data 2 and compared with the wild-type 3D232 using DNAMAN 8 software (Lynnon Biosoft, San Ramon, CA, USA).
Wheat transformation
The coding sequence (CDS) of the CYB561-Domon and TM9SF4 genes were amplified from 3D232 using the primers listed in Supplementary Data 2 and inserted downstream of the maize (Zea mays L.) Ubiquitin (Ubi) promoter in a pTCK303 vector digested by Kpn I and Spe I to generate the overexpression vector ProUbi: CYB561-Domon and ProUbi:TM9SF4, respectively. A 10,741 bp genomic DNA fragment that contains the 3277 bp full-length WTK7-TM coding region, as well as 3311 bp upstream of the start codon and 4,153 bp downstream of the stop codon, was cloned into the vector pCAMBIA1300 to construct ProWTK7-TM: WTK7-TM for transformation. These resulting plasmids were transformed into the Agrobacterium tumefaciens strain EHA105 and delivered into the soft white spring wheat cultivar Fielder by the A. tumefaciens-mediated transformation method, respectively65. Fifteen individuals of T1 transgenic families were inoculated with Bgt isolate E09. The transgenic plants were identified by PCR amplification and sequencing. The gene expression was measured with qRT-PCR as mentioned above. TaActin was used as an endogenous control. Expression values are means ± SD from three biological replicates (three leaves used per biological replicate) and three technical replicates for each leave. All primers used for the construction preparation, the transgenic plant identification, and gene expression are listed in Supplementary Data 2.
Subcellular localization assay
For subcellular localization, the coding sequence of WTK7-TM was inserted into the binary vector pCAMBIA1300-GFP driven by the 35 S promoter to construct the recombinant plasmid 35 S:: WTK7-TM-GFP. Arabidopsis NAA60 was used as a plasma membrane marker protein described by Linster et al.66. Fusion proteins 35 S::WTK7-TM-GFP + 35 S::NAA60-mCherry or 35 S::GFP + 35 S::NAA60-mCherry were expressed in N. benthamiana leaves through agroinfiltration. For plasmolysis experiments, leaf samples were incubated for 10–15 min in 4% NaCl before imaging45. N. benthamiana leaves were imaged 2dpi using a confocal laser scanning microscope (Carl Zeiss, LSM880).
Protein extraction and analysis
N. benthamiana leaves were frozen in liquid nitrogen immediately after harvest. Subsequently, the leaves were finely ground into powder and transferred into 1.5 mL centrifuge tubes. Total proteins were extracted using lysis buffer (10 mM Tris-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 10 mM DTT, 1×complete protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride (PMSF)). After vortexing for 20 min at 4 °C, the samples were centrifuged at 14,000 × g for 10 min at 4 °C. Then transferred the supernatant to a new 1.5 mL Eppendorf tube. The protein samples were boiled with 1×SDS loading buffer and analyzed by immunoblotting. The proteins were detected by SDS-PAGE using Anti-GFP pAb-HRP-DirecT (MBL, 1:3000 dilution) antibodies.
In vitro phosphorylation
For the in vitro phosphorylation assay, the indicated lengths of WTK7-TM CDS were cloned into the vector pMAL-c5x to obtain MBP-WTK7-TMKin I, MBP-WTK7-TMKin II, MBP-WTK7-TMKin I-Kin II and MBP-WTK7-TM-FL constructs. Recombinant proteins were induced in DE3 (Rosetta) strain with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 12 h at 16 °C. 100 mL cell culture was collected at 4 °C by centrifugation and the pellet was resuspended in 20 mL PBS buffer (200 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, pH 7.4) with 1 mM PMSF, 5 mM DTT and 0.05% Triton-X 100. The suspension was sonicated for 8 min and centrifugated at 8000 × g for 25 min. Then the supernatant was incubated with amylose resin for 2–3 h at 4 °C and MBP-tagged proteins were eluted with 10 mM maltose in PBS buffer. The purified proteins were separated by 10% SDS-PAGE and stained with coomassie brilliant blue to examine the purity. 5 μg indicated MBP-tagged proteins were incubated with 2 μg myelin basic protein in kinase reaction buffer (50 mM Hepes (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.1 mM ATP, 1 μCi [γ-32P]-ATP) for 20 μL volume. After being kept in 30 °C for 30 min, 5×SDS loading buffer was added into the mixture to stop the reaction and the proteins were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The radioactivity was detected with the Typhoon FLA 9500 (GE Healthcare Life Sciences, Piscataway, USA).
Protein sequence and domain analysis
The protein sequence of WTK7-TM was predicted on the basis of the conserved domain database from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART. Prediction of transmembrane helices was performed with TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/), Phobious (https://phobius.sbc.su.se/) and Protter (http://wlab.ethz.ch/protter/#). The three-dimensional structure of WTK7-TM was predicted using AlphaFold v2.039. The structural graphic was generated using PyMOL (The PyMOL Molecular Graphics System, v.2.0; Schrödinger).
Phylogenetic analysis
Phylogenetic analysis was conducted to compare putative kinase domains including 184 putative kinase or pseudokinase domains used in the study ofWTK124, and 12 putative kinase or pseudokinase domains of WTK225, WTK315, WTK428, WTK5 (Sr62)26, WTK6-vWA (Lr9)27, and WTK7-TM. ClustalW in the MEGA was used for multiple alignment using the default setting and phylogenetic construction. Phylogenetic trees were built with the Neighbor-joining method in the MEGA and polished by Itol (https://itol.embl.de/).
Functional marker development
Based on the genomic sequence of the WTK7-TM gene, we developed a specific primer, WTK7-TM-FM, to identify the presence of the WTK7-TM functional allele among Triticeae species. The dominant functional marker WTK7-TM-FM amplified a 1198 bp PCR product if the plant carrying WTK7-TM gene, otherwise no amplification product. A 2 × Taq Master Mix (Vazyme, China) was used in a total volume of 10 µL with the following amplification program: 95 °C for 3 min, 40 cycles of 95 °C for 15 s, 60 °C for 15 s and 72 °C for 30 s, 72 °C for 10 min. PCR products of functional marker WTK7-TM-FM were mixed with 2 μL of loading buffer (98% formamide, 10 mM EDTA, 0.25% bromophenol blue, and 0.25% xylene cyanol) and separated on 1% agarose gels. The coding sequence (CDS) of the WTK7-TM gene was amplified from 38 WEW accessions, which were tested carrying WTK7-TM gene by functional marker WTK7-TM-FM, using the primers listed in Supplementary Data 2. ClustalW was used for multiple sequence alignment.
Primers
Primers used in this study are listed in Supplementary Data 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings of this work are available within the paper and its Supplementary Information files. Transcriptome data generated from the current study are publicly available from the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Bei**g Institute of Genomics, Chinese Academy of Sciences at the accession CRA009664. The sequence of Pm36 was deposited in the National Center for Biotechnology Information (NCBI) under the accession number OQ361691. Source data are provided in this paper.
References
Goel, S. et al. Exploring diverse wheat germplasm for novel alleles in HMW-GS for bread quality improvement. J. Food. Sci. Technol. 55, 3257–3262 (2018).
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
Cowger, C. et al. Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology 108, 402–411 (2018).
Hewitt, T. et al. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826 (2021).
Sánchez-Martín, J. et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221 (2016).
Yahiaoui, N., Srichumpa, P., Dudler, R. & Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37, 528–538 (2004).
Sánchez-Martín, J. et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 7, 327–341 (2021).
**e, J. et al. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228, 1011–1026 (2020).
Hurni, S. et al. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969 (2013).
Zhu, S. et al. Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 4, 100472 (2023).
Li, H. et al. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat. Commun. 15, 2449 (2024).
Singh, S. P. et al. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98, 249–260 (2018).
He, H. et al. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882 (2018).
**ng, L. et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878 (2018).
Lu, P. et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680 (2020).
Krattinger, S. G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009).
Li, M. et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228, 1027–1037 (2020).
Moore, J. W. et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498 (2015).
Zhao, Y. et al. Pm57 from Aegilops searsii encodes a novel tandem kinase protein conferring powdery mildew resistance in bread wheat. Preprint at Research Square https://doi.org/10.21203/rs.3.rs-2844708/v1 (2023).
Zou, S., Wang, H., Li, Y., Kong, Z. & Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 218, 298–309 (2018).
Li, Y. et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 5, 100646 (2024).
Gaurav, K. et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 40, 422–431 (2022).
Klymiuk, V. et al. Tandem protein kinases emerge as new regulators of plant immunity. Mol. Plant Microbe. Interact. 34, 1094–1102 (2021).
Brueggeman, R. et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99, 9328–9333 (2002).
Klymiuk, V. et al. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9, 3735 (2018).
Chen, S. et al. Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 225, 948–959 (2020).
Yu, G. et al. Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nat. Commun. 13, 1607 (2022).
Wang, Y. et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 55, 914–920 (2023).
Arora, S. et al. A wheat kinase and immune receptor form host-specificity barriers against the blast fungus. Nat Plants 9, 385–392 (2023).
Blanco, A. et al. Molecular map** of the novel powdery mildew resistance gene Pm36 introgressed from Triticum turgidum var. dicoccoides in durum wheat. Theor. Appl. Genet. 117, 135–142 (2008).
Zhang, H. et al. Genetic and comparative genomics map** reveals that a powdery mildew resistance gene Ml3D232 originating from wild emmer co-segregates with an NBS-LRR analog in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 121, 1613–1621 (2010).
Zhang, D. et al. Comparative genetic map** revealed powdery mildew resistance gene MlWE4 derived from wild emmer is located in same genomic region of Pm36 and Ml3D232 on chromosome 5BL. J. Integr. Agric. 14, 603–609 (2015).
Avni, R. et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97 (2017).
Maccaferri, M. et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895 (2019).
International Wheat Genome Sequencing Consortium, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar 7191 (2018).
Walkowiak, S. et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283 (2020).
Sato, K. et al. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res. 28, 1–7 (2021).
Yuan, C. et al. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6, e26468 (2011).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Ngou, B. P. M., Ding, P. & Jones, J. D. G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478 (2022).
Couto, D. & Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 16, 537–552 (2016).
Wang, W., Wen, Y., Berkey, R. & **ao, S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898–2913 (2009).
Dinh, H. X. et al. The barley leaf rust resistance gene Rph3 encodes a predicted membrane protein and is induced upon infection by avirulent pathotypes of Puccinia hordei. Nat. Commun. 13, 2386 (2022).
Büschges, R. et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705 (1997).
Kolodziej, M. C. et al. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nat. Commun. 12, 956 (2021).
Saintenac, C. et al. A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against Septoria tritici blotch. Nat. Commun. 12, 433 (2021).
Alkan, C., Coe, B. & Eichler, E. Genome structural variation discovery and genoty**. Nat. Rev. Genet. 12, 363–376 (2011).
Chen, Z. et al. Structural variation at the maize WUSCHEL locus alters stem cell organization in inflorescences. Nat. Commun. 12, 2378 (2021).
Athiyannan, N. et al. Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nat. Genet. 54, 227–231 (2022).
Meyer, R. & Purugganan, M. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013).
Huang, L. et al. Distribution and haplotype diversity of WKS resistance genes in wild emmer wheat natural populations. Theor. Appl. Genet. 29, 921–934 (2016).
Chen, P. et al. Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breeding 31, 477–484 (2013).
Hou, J. et al. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 164, 1918–1929 (2014).
Gollner, K., Schweizer, P., Bai, Y. & Panstruga, R. Natural genetic resources of Arabidopsis thaliana reveal a high prevalence and unexpected phenotypic plasticity of RPW8-mediated powdery mildew resistance. New Phytol. 177, 725–742 (2008).
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A. & Allard, R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8018 (1984).
Kosambi, D. D. The estimation of map distances from recombination values. Ann. Hum. Genet. 12, 172–175 (1943).
Mayjonade, B. et al. Extraction of high-molecular-weight genomic DNA for long-read sequencing of single molecules. BioTechniques 61, 203–205 (2016).
Travers, K. J. et al. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids. Res. 38, e159 (2010).
Cheng, H. et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Solovyev, V., Kosarev, P., Seledsov, I. & Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, S10.1–S10.12 (2006).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Kim, D. et al. Graph-based genome alignment and genoty** with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Saintenac, C. et al. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341, 783–786 (2013).
Ishida, Y., Tsunashima, M., Hiei, Y. & Komari, T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198 (2015).
Linster, E. et al. The Arabidopsis Nα-acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response. New Phytol. 228, 554–569 (2020).
Acknowledgements
We are grateful to Profs. Qixin Sun and Tsomin Yang, China Agricultural University, for their advices and support during the research. Many thanks to Drs. Xueyong Zhang, **anchun **a, and Lihui Li of the Chinese Academy of Agricultural Sciences, Bei**g, China for providing the Chinese wheat mini-core collection (MCC), worldwide wheat landrace collection, and Chinese wheat landraces, respectively; Dr. Antonio Blanco of the University of Bari Aldo Moro, Bari, Italy for providing resistant line 5BIL-29. This research was financially supported by the National Key Research and Development Program of China (2021YFA1300703 to Z.Y.L.), National Natural Science Foundation of China (32101735 to M.M.L. and U21A20224 to Z.Y.L.), Key Research and Development Program of Hebei Province (22326305D to C.G.Y) and Hainan Seed Industry Laboratory (B21HJ0111 to Z.Y.L.).
Author information
Authors and Affiliations
Contributions
Z.L., M.L. and Y.Z. designed the study. M.L., H.Z., H.X., K.Z., W.S., D.Z., Y.W., L.Y., Q.W., Y.C., D.Q., P.L., B.L., L.D., W.L., X.C., L.L., X.T. and C.Y. performed the experiments, conducted fieldwork, analyzed data, performed Bgt inoculation, and developed EMS mutants. L.L.D., J.X. and G.G. performed bioinformatic analysis. Y.L., D.Y., E.N., T.F. and H.L. provided powdery mildew resistance phenoty**, germplasm, or scientific support and advice. Z.L., M.L. and Y.Z. wrote the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Manuel Spannagl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Zhang, H., **ao, H. et al. A membrane associated tandem kinase from wild emmer wheat confers broad-spectrum resistance to powdery mildew. Nat Commun 15, 3124 (2024). https://doi.org/10.1038/s41467-024-47497-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-47497-w
- Springer Nature Limited
This article is cited by
-
Pm57 from Aegilops searsii encodes a tandem kinase protein and confers wheat powdery mildew resistance
Nature Communications (2024)