Abstract
Mesoscopic calcium imaging enables studies of cell-type specific neural activity over large areas. A growing body of literature suggests that neural activity can be different when animals are free to move compared to when they are restrained. Unfortunately, existing systems for imaging calcium dynamics over large areas in non-human primates (NHPs) are table-top devices that require restraint of the animal’s head. Here, we demonstrate an imaging device capable of imaging mesoscale calcium activity in a head-unrestrained male non-human primate. We successfully miniaturize our system by replacing lenses with an optical mask and computational algorithms. The resulting lensless microscope can fit comfortably on an NHP, allowing its head to move freely while imaging. We are able to measure orientation columns maps over a 20 mm2 field-of-view in a head-unrestrained macaque. Our work establishes mesoscopic imaging using a lensless microscope as a powerful approach for studying neural activity under more naturalistic conditions.
Similar content being viewed by others
Introduction
Mesoscopic imaging provides an opportunity to discover how behaviors are represented by large-scale brain activity patterns that occur over multiple brain areas. The development and application of mesoscopic imaging effectively balances the need for high spatiotemporal resolution recording with remarkably large fields-of-view (FOVs). This technology has been demonstrated through its application in multiple species including rodents1,2,3,4, cats5,6,7,8,9 and primates10,11,12,13,14,15. When applied to rhesus macaques, mesoscale imaging can help uncover cognitively sophisticated behaviors like perception, motor planning and decision-making because these non-human primates (NHPs) have close genetic, anatomical and behavioral similarity to humans16,17,18. Imaging provides unique advantages for researchers to non-invasively record high density neural activity from large neuron populations13,19,20,21. For example, one can study perception in the primary visual cortex (V1). Specifically, primates display pinwheel-like structures that segregate neurons into columns based on edge orientation with periodicity of ~1.2 cycles/mm15,22,23,24,25, while rodents have a random distribution of orientation-selective neurons, resulting in salt-and-pepper organization24,26. The success of these efforts can be attributed to the refinement of surgical procedure, and improvement of genetically encoded calcium indicators (GCaMP)27,28 and the associated viral preparation13,29,30,31,32,33,34,68 as well as a finer-scale orientation map7,44,66. We first focused on the retinotopic map. When performing the experiments, we selected a visual stimulus pattern that has been reported to elicit reliable position-tuning curves in macaque V113,25,66. Specifically, we used a small flashed grating as the visual stimulus with 0.25 degree stimulus size and 4 cycle per degree (cpd) spatial frequency presented at different locations in the visual field. Stimulus position vertical coordinates in degree of visual angle varied from −0.8 degree to −1.3 degree, with a change of 0.1 degree change (Fig. 3c). During the experiments, the macaque performed a visual fixation task, kee** fixation within a window of less than 2 degree width, centered on a small fixation point for 2–4 s. An infrared eye tracker (EyeLink) was used to monitor the animal’s eye position. A second identical grating was flashed at a mirror-symmetric location in the opposite hemifield to help the macaque maintain eye fixation. The stimuli were flashed 4 times per trial at 4 Hz frequency (100 ms ON and 150 ms OFF). All stimulus conditions were randomly interleaved and repeated for 10 times each, and the stimulus trials were mixed with blank fixation trials. The recorded videos were captured at 20 Hz for both Bio-FlatScopeNHP and widefield microscope.
From the Bio-FlatScopeNHP reconstructions, it is clear that GCaMP signals were entrained with each stimulus cycle suggesting that we were indeed recording neural activity with the Bio-FlatScopeNHP (Fig. 3b). We first registered the Bio-FlatScopeNHP reconstructions with those from widefield microscopy based on blood vessel structures. From the reconstructions of the response to 6 stimuli at different positions, it is evident that the active areas captured by both devices show good correspondence (Fig. 3f). We selected six regions of interest (ROIs) for tuning curves analysis, with a size of ~0.09 mm2 each and the distance of around 0.37 mm between adjacent ROIs (Fig. 3d, e). We recorded 2 sessions of Bio-FlatScopeNHP experiments (additional results in Supplementary Fig 10) and 1 session of a widefield microscope experiment. The Gaussian fitted tuning curve for the 6 ROIs shows excellent agreement between Bio-FlatScopeNHP reconstructions and widefield microscope captures (Fig. 3d, e). The peak position of each fitted tuning curve indicates the corresponding visual location at each ROI, which shows a difference between two devices of less than 0.031 degree (average difference = 0.014 degree) (Fig. 3g).
Despite the high level of agreement between Bio-FlatScopeNHP reconstruction and widefield microscope captures, we noticed a lower ∆F/F in Bio-FlatScopeNHP reconstructions compared to the widefield microscope, especially at the edge of the FOV. One possible explanation could be the lower signal-to-noise ratio in Bio-FlatScopeNHP reconstructions, and the illumination profile can also potentially affect the reconstruction quality (Supplementary Fig 7). Nevertheless, this experiment confirmed that Bio-FlatScopeNHP can capture calcium dynamics and obtain high-quality position-tuning information that is comparable to traditional table-top widefield microscopes.
In vivo calcium imaging of orientation tuning maps in a head-fixed macaque V1
We further found that Bio-FlatScopeNHP is capable of capturing calcium dynamics at a columnar scale comparable to a table-top widefield microscope system. Obtaining high-quality orientation maps is more challenging than position-tuning curves due to the relatively small spatial scale and weak orientation-selective signals.
In this experiment, we used grating visual stimuli that have been previously reported to reliably elicit orientation selective signals in macaque V112,13,66. While the macaque maintained a steady gaze at the center of the screen, large high contrast sinusoidal grating stimuli at 6 equally spaced orientations (0, 30, 60, 90, 120, 150 degrees) were flashed at a frequency of 4 Hz for 4 cycles, with a spatial frequency of 4 cpd. A second identical grating was flashed at a mirror-symmetric location in the opposite hemifield to help the macaque maintain eye fixation. The size of the grating is 6 deg x 6 deg, which is large enough to cover the whole imaging area. All stimulus conditions were randomly interleaved, repeated for 10 times each, and mixed with blank fixation trials. We recorded 2 sessions of Bio-FlatScopeNHP experiments (additional results in Supplementary Fig. 11) and 2 sessions of widefield microscope experiments.
We obtained precise orientation maps from the Bio-FlatScopeNHP reconstructions and observed strong GCaMP signals during each stimulus cycle (Fig. 4b) with ∆F/F levels comparable to those captured by the widefield microscope. We first registered the Bio-FlatScopeNHP reconstructions with those from the widefield microscope using blood vessel structures as a reference. Using root-mean-square (RMS) maps calculated on averaged amplitude of the orientation tuning at each pixel location (Supplementary Fig 9), we selected ROIs with strongest 4 Hz signals (Fig. 4c). To analyze the orientation maps from the captured signals, we computed the 4 Hz Fourier amplitude of the average GCaMP signal at each location, and applied a bandpass spatial filter (0.8–2.5 cycle/mm) to remove non-orientation-selective responses and high-frequency noise (Fig. 4f). We employed a linear orientation decoder69 to quantitatively evaluate the orientation discriminability of the captured columnar-scale neural responses, as follows. We first calculated pixel-wise d’ maps from the set of single-trial 0 degree and 90 degree orientation maps, where each pixel showed the d’ for discriminating between the two orientations at that pixel. These d’ maps were then used as linear weights to compute a decision variable per trial for each orientation (Fig. 4e). The separability of these decision variables between the two orientations was used to compute an overall d’ (see Methods). The decoder uses pixel-wise summation of single-trial responses, weighted by corresponding d’ maps, and summed over the selected ROI (see Method). With cross-validation (see Method), we calculated the discriminability of the pooled signals in distinguishing between 0 degree and 90 degree orientations, which can be interpreted as the signal-to-noise ratio of the decoder. We found a high discriminability (d’) in both the ground truth captures (d’ = 29.90) and Bio-FlatScopeNHP reconstructions (d’ = 6.58) (Fig. 4e). Representative maps of 0 degree and 90 degree are shown in Fig. 4d. To verify the validity of the observed map, we computed the Pearson correlation between each pair of maps, and averaged the correlations across all pairs of maps with the same stimulus orientation difference (Fig. 4g). As the difference in orientation between two maps increases, their correlations follow a systematic pattern. When the two orientations differ by 45 degrees, the correlation approaches zero, and for an orientation difference of 90 degrees, the correlation reaches a maximum negative value of −0.81 for ground truth and −0.75 for Bio-FlatScopeNHP, which matched the known structure of orientation maps in V112,13,66. We then used the maps at each orientation to compute the composite orientation map (Fig. 4h), in which the color indicates the preferred orientation and saturation the strength of orientation tuning. This composite map reveals the semi-periodic organization of the orientation map, including the orientation pinwheels7,69. To assess the accuracy of Bio-FlatScopeNHP captures, we compared the composite orientation maps obtained from Bio-FlatScopeNHP and the widefield microscope by calculating the correlation coefficient. We converted the color map of preferred orientation to grayscale map in the range of [−1 1] by taking the sine of the preferred orientation times two13. We computed the correlation coefficients between the converted maps in each pixel from the maps calculated from Bio-FlatScopeNHP and those obtained using the widefield microscope. The correlation coefficients between the maps calculated from Bio-FlatScopeNHP and the widefield microscope were found to be similar (correlation coefficients = 0.87 and 0.85) as compared to the correlation coefficient between two sessions captured by the widefield microscope (correlation coefficient = 0.92). This experiment demonstrated that Bio-FlatScopeNHP is capable of capturing calcium dynamics at columnar scale and producing high quality orientation map information that is comparable to traditional table-top widefield microscopes.
a Illustration of experimental setups for capturing ground truth (left) and Bio-FlatScopeNHP imaging (right). In the experiments, flashed large gratings at six different orientations are used as visual stimuli. b Average time course of GCaMP response to the flashed gratings from ground truth (top) and Bio-FlatScopeNHP reconstructions (bottom). ∆F/F indicates the relative changes in fluorescence. Shaded area ± SEM. c Left: Example images captured by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). Scale bar, 500 μm. The yellow square indicates the selected overlap** ROI. d Response maps of orientation-selective GCaMP signals obtained by bandpass filtering the response maps to 0-degree and 90-degree orientations (out of six evenly spaced orientations) obtained by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). e Decision variables calculated from 0-degree and 90-degree trials captured by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). f Spatial 1D amplitude spectrum of the single-orientation response maps obtained by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). The spatial filtration removes components outside spatial frequency (SF) between 0.8 and 2.5 cycles/mm, indicated by the shaded area. g Pairwise correlations between all six orientation maps as a function of stimulus orientation difference obtained by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). Shaded area ± SEM. h Orientation maps obtained by ground truth widefield microscope (top) and Bio-FlatScopeNHP (bottom). Scale bar, 500 μm. Source data are provided as a Source Data file.
In vivo calcium imaging of orientation tuning maps in a head-unrestrained macaque V1
Since the Bio-FlatScopeNHP produces images of similar quality to those captured by the table-top microscope system for head-fixed macaques, we were able to release the head fixation and achieve an important milestone - imaging columnar-scale neural activity in a head-unrestrained macaque.
By imaging a head-unrestrained macaque, we demonstrated that Bio-FlatScopeNHP can successfully capture calcium dynamics at a columnar scale even when the animal’s head is unrestrained. The most significant advantage of Bio-FlatScopeNHP compared to traditional table-top systems is its small form factor, which allows it to be mounted directly on the intracranial implant. In this experiment, we took advantage of this feature and imaged the orientation tuning properties of V1 GCaMP response across different stimulus orientations on a head-unrestrained macaque. In the same experimental session, we also recorded a head-fixed block of trial using the widefield microscope and a head-fixed Bio-FlatScopeNHP block (Supplementary Fig 12) for comparison, all within a two-hour time window. Same visual stimulus with 6 different orientations was used in the widefield microscopic session. For the head-unrestrained experiment, we moved the visual stimulus to half distance (54 cm) and enlarged the visual stimulus pattern to 20 degree stimulus size and 3 cpd spatial frequency presented at different orientations (Fig. 5a). Enlarging the stimulus size alleviates the requirement for strict eye fixation under head-unrestrained conditions. The subject was trained to look at the fixation point at the center of the screen after an audio cue to get a reward. However, in contrast to the head-fixed condition, we did not enforce fixation in the head-unrestrained trials.
a Illustration of experimental setups for head-fixed imaging (left) and head-unrestrained imaging (right). In the experiments, large gratings at six different orientations are used as visual stimuli. During head-unrestrained experiments, visual stimuli were moved closer to the animal and the stimulus was larger and at a higher spatial frequency. b Average time course of GCaMP response to the flashed gratings from head-fixed imaging (left) and head-unrestrained imaging (right). ∆F/F indicates the relative changes in fluorescence. Shaded area ± SEM. c Example images captured by widefield microscope (top) in a head-fixed imaging session and Bio-FlatScopeNHP (bottom) in a head-unrestrained session. Scale bar, 500 μm. The yellow square indicates the selected overlap** ROI. d Spatial 1D amplitude spectrum of the single-orientation response maps obtained by head-fixed imaging (top) and head-unrestrained imaging (bottom). The spatial filtration removes components outside spatial frequency (SF) between 0.8 and 2.5 cycles/mm, indicated by the shaded area. e Pairwise correlations between all six orientation maps as a function of stimulus orientation difference obtained by head-fixed imaging (top) and head-unrestrained imaging (bottom). Shaded area ± SEM. f Orientation maps obtained by head-fixed imaging (top) and head-unrestrained imaging (bottom). Scale bar, 500 μm. Source data are provided as a Source Data file.
We were able to obtain an accurate orientation map from the Bio-FlatScopeNHP reconstructions on the head-unrestrained macaque and observed clear GCaMP signals during each stimulus cycle (Fig. 5c) with a decreased ∆F/F compared to the head-fixed captures. We registered the reconstructions from head-unrestrained sessions with those from the widefield microscope captures based on blood vessel structures. We used the same RMS maps to select the region of interest as in previous experiments, and applied the same bandpass spatial filter (0.8–2.5 cycle/mm) on the 4 Hz Fourier amplitude of the averaged GCaMP signal to remove non-orientation-selective responses and high frequency noise (Fig. 4d). To verify that observed maps are actual orientation columns, we computed the Pearson correlation between each pair of maps, and averaged the correlations across all pairs of maps with the same stimulus orientation difference (Fig. 5e). The pairwise correlations obtained by Bio-FlatScopeNHP from the head-unrestrained animal still smoothly change from positive values for maps produced by nearby orientations to negative values for maps produced by orthogonal orientations, reaching a maximum negative value of −0.81 for head-fixed session and −0.54 for head-unrestrained session (Fig. 5e). We calculated the map captured from Bio-FlatScopeNHP imaging of the head-unrestrained animal, and compared it to the map obtained from a head-fixed animal using the widefield microscope (Fig. 5f). The map from the head-unrestrained animal still revealed the semi-periodic organization of the orientation map, and the orientation pinwheels44 are still visible but with blurrier edges. To evaluate how closely the head-unrestrained orientation maps matched the head-fixed condition, we computed the correlation to random orientation maps by randomly labeling the 6 orientations from the head-unrestrained animal 1500 times and calculating the correlation coefficient. The maps generated from the shuffled data showed random correlation coefficients ranging from −0.08 to 0.48 (average = 0.25). The actual correlation coefficient between the head-unrestrained animal and the head-fixed animal is 0.72, indicating that these maps show significant similarities that are not explained by chance (Supplementary Fig. 14).
Discussion
In this study, we present a lensless microscope capable of imaging large FOVs (over 20 mm2) at high resolution in the primary visual cortex of the macaque. The small and light form factor allows us to perform widefield in vivo calcium imaging on a head-fixed awake macaque from which we were able to extract position tuning properties and orientation maps. These maps showed good correspondence with those captured by a traditional table-top widefield microscope. A key advantage of head-unrestrained imaging is the ability to study eye-head movement coordination during natural behaviors. During natural behaviors, the head and eyes move simultaneously and in a coordinated fashion. Because it has been previously impossible to imaging large-scale neural activity during this complex behavior, many open questions remain regarding the neural mechanisms that mediate the coordination of the head and eyes and the coordinate transformations that are necessary to guide motor responses based on multimodal sensory evidence. Our technology would allow researchers to study how sensory areas, such as V1 and auditory cortex70,71, and sensory motor areas72,73, such as the premotor cortex and frontal eye fields, are affected by natural head and eye movements. Another area where the Bio-FlatScopeNHP could play an important role is understanding social interactions74,75 and emotions76 in primates. Macaques are highly social animals and recent studies have started to look at the neural basis of social interactions between NHPs. To better understand social behavior and its underlying neural representations, it is essential to study behavior and neural representations in head unrestrained animals. While many of these questions may be addressable with head-unrestrained animals, some of these questions would be best studied in freely behaving animals. To facilitate studies in freely behaving animals, future versions of the Bio-FlatScopeNHP should be made wireless, protected in a more robust housing, and more robust hemodynamic correction should be added. In this proof-of-principle study in a head-unrestrained animal, we used heartbeat-triggered trials and a fast, periodic stimulus presentation to overcome hemodynamic artifacts and improve SNR. For future continuous recording under more naturalistic conditions, one could compensate for the hemodynamic artifacts by recording the reflectance signal using a color that does not interfere with the fluorescence channel, as has been used in rodent imaging. For example, GCaMP imaging in mice can achieve hemodynamic correction by using a color camera to record the reflectance signal of green and red LEDs2,4,41. Future versions of the Bio-FlatScopeNHP could use a similar approach by changing the emission filter. Alternatively, future versions of the Bio-FlatScope NHP could use interleaved blue and green illumination with a trigger circuit to image fluorescence and reflectance signal simultaneously41, which has been shown in rodent imaging.
One potential limitation of our approach is that the overall signal-to-noise ratio (SNR) in Bio-FlatScopeNHP reconstructions is lower compared to traditional table-top systems, particularly at the edge of the FOV. This can lead to a decrease in ∆F/F compared to the widefield microscope imaging. This could be attributed to uneven illumination at the edges of the FOV. However, this issue can be improved by optimizing the phase mask and illumination setups to allow imaging at closer distance at better illumination conditions. Imaging at closer distance can increase the light collection efficiency and improve the system resolution and SNR. Another contributing factor is that the imaging experiments in this study were conducted approximately 1.5 years post injection, which could have led to a reduction in signal levels. When imaging is performed on a subject that has recently received the injection, it is likely that we could achieve higher signal levels and an improved SNR.
An additional constraint limiting the size of our in vivo FOV is the dimensions of the chamber insert. Our illumination design is restricted by the small size (21 mm diameter) and the large depth (8 mm) of the chamber insert. When removing this constraint, our device demonstrates the capability to achieve a significantly expanded FOV (64 mm2), as shown in Supplementary Fig 4 and Supplementary Fig 7d. Compared to current head-mounted devices used in rodents37,38,41,49,59,77,78,79,80, the Bio-FlatScopeNHP stands out as the only system that combines a peak resolution of < 10 μm and a FOV over 60 mm2 (Fig. 1f). Although our system demonstrated a < 10 μm resolution on test target, individual fluorescent cells in vitro, and NHP blood vessels roughly 20 μm in diameter (Supplementary Fig 5e), the dense GCaMP labeling and scattering by the brain tissue prevents single-cell resolution fluorescent imaging in vivo. This is also true for wide-field imaging with similar dense labeling, where individual cells cannot be resolved. Given the < 10 μm optical resolution of the Bio-FlatScopeNHP we expect that single-cell resolution imaging may be possible when sparse labeling strategies developed in mice81 are translated to NHPs. These strategies include using cell-type specific promoters to label sparse neural populations such as subclasses of inhibitory neurons82,83, combining retrograde AAV injections in downstream regions84,85 together with intersectional approaches such as Cre or Flp82 to strongly label sparse populations of projections cells from superficial layers, and using genetic motifs that restrict GECI expression to the soma.
While we demonstrate the Bio-FlatScopeNHP in rhesus macaque, the lightweight design should be compatible with a number of other common animal models. The largest contributor to the weight of our current prototype (weight ~17.2 g) is our off-the-shelf CMOS sensor (~4.9 g) and 3D printed housing (~8.6 g) used for mounting on the cortex chamber. We compared the weight of the Bio-FlatScopeNHP with head-mounted devices currently utilized in several other animal models, including rats59, ferrets60 and common marmosets61 (Supplementary Fig 1). Notably, the overall weight of the Bio-FlatScopeNHP is lighter than the weight that can be carried on these animals, which indicates the potential for in vivo imaging studies across species. We envision our device as a promising candidate for conducting in vivo imaging with large FOVs on marmosets - an emerging NHP model system in neuroscience. Marmosets have shallower cortical surfaces compared to macaques, offering the possibility of imaging directly from outside the cortex chamber. This releases the constraints on illumination design posed by current chamber insert sizes, allowing our system to achieve an even larger FOV in vivo (Supplementary Fig 7). An additional challenge of imaging marmoset brains is the smaller size of the brain compared to macaques. The curved nature of the brain surface can potentially impact imaging quality, where lens-based systems have limited depth of focus. One distinct advantage of our system compared to current lens-based devices is the capability to perform digital refocusing. By using point spread functions obtained at different depths, this digital refocusing mechanism enables us to bring areas that are out of focus back into focus (as shown in Supplementary Fig 5e). This unique refocusing ability is a key advantage for imaging over curved surfaces like the surface of the marmoset brain. To facilitate use in even smaller animals or the use of multiple microscopes over different brain areas in the same animal, future designs can be made smaller and lighter by using a small and lightweight sensor printed circuit board (PCB) and electronics, as well as lightweight 3D printed materials for the housing. Additionally, incorporating miniaturized OLEDs for on-chip illumination can significantly reduce the form factor of the device and facilitate its integration, potentially leading to the development of an implantable device86,87.
Overall, in this paper, we have demonstrated that Bio-FlatScopeNHP can capture calcium dynamics in macaque V1 at the columnar scale and provide high-quality orientation map information from a head-unrestrained NHP, opening a path for studying brain activity in these animals under more naturalistic conditions.
Methods
Device fabrication
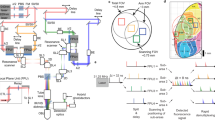
The prototype was constructed utilizing a commercially available board level camera (Imaging Source, DMM 37UX187-ML) with a monochrome Sony CMOS imaging sensor (IMX178LLJ with 6.3 MP and 2.4 μm pixels). The prototype employed a 3.5 mm sensor-to-mask distance and an approximate 3 mm working distance for fixed samples and in vivo imaging.
The PSF pattern for the phase mask was generated by using Canny edge detection on a randomly generated Perlin noise (Supplementary Fig 3) with a feature size of 6 μm, which was selected based on the fabrication limit. The phase mask was then designed using a phase retrieval algorithm58 using the PSF pattern. The phase mask was fabricated using a 3D maskless two-photon photolithography system (Nanoscribe, Photonic Professional GT) in high-resolution dip-in liquid lithography mode. The mask was fabricated on a 700 μm thick fused silica substrate using a photoresist (IP-Dip). The laser power used for fabrication was 65% of the maximum power, and it should be adjusted based on systems for optimal results by clear visualization in the real-time monitoring software. After exposure, the fabricated mask was immersed in SU-8 developer for 20 min, followed by a 2 min soak in isopropyl alcohol. The size of the fabricated phase mask is 2.3 mm × 2.3 mm and has a height of 1 μm with a 200 nm step and a 1 μm fabrication pixel size. The substrate was laser cut to 7 mm × 7 mm to fit the design of the housing and fitted with an opaque mask to create an aperture containing only the phase mask. A hybrid filter set49,62 was attached after the phase mask, consisting of a commercially available adsorption filter (Kodak, Wratten 12) and a custom-designed interference filter (Chroma, ET525/50 m) with a thickness of 500 μm. The filters and phase mask were housed in a 3D printed enclosure (printed using Formlabs Form 3).
The integrated illumination system consisted of four surface-mounted LEDs (LXML-PB01-0030) with excitation filters (Chroma, ET470/40x). The LEDs were symmetrically positioned around the imaging module and angled at 40 degrees to direct light towards the central FOV (Supplementary Fig 7). The excitation power measured at the 3 mm working distance was approximately 0.3 ~ 1.0 mW/mm2, which is comparable to that of the table-top widefield system (0.1 ~ 0.2 mW/mm2) and sufficient for one-photon calcium imaging in the macaque cortex. The entire device was mounted on the cortex chamber using a 3D-printed holder (printed using ProJet MJP 2500).
Device calibration
A one-time calibration procedure must be completed to record the experimental PSF of the mask prior to conducting imaging experiments. The point source we used for calibration was a 10 μm pinhole that was illuminated by a green LED (Thorlabs, M530L4) placed behind an 80-degree holographic diffuser. Images of the central PSFs were obtained at 20 μm intervals across a working distance range of 0.5 mm–6 mm. These central PSFs were used in fast reconstructions using a shift-invariant deconvolution model to assess the positioning of the device quality of the imaging. For the shift-variant deconvolution model, calibration was performed at 20 μm intervals within a working distance range of 2.9 mm−3.1 mm. At a specific depth, the distance between each calibration measurement was 1 mm, and a 9 × 9 grid was calibrated (81 PSFs for one depth) on the imaging plane, covering an imaging area of 8 mm × 8 mm. The calibrations were performed automatically by using programmable motorized linear translation stages (Thorlabs, LNR502). All calibration images were averaged through five captured images to improve the signal-to-noise ratio. Calibration across a range of working distances enables digital refocusing of the images in post processing.
Image reconstruction
Image reconstruction is a crucial process for lensless microscopes and is commonly approached as a convex optimization problem. To achieve fast reconstruction using single PSF captured at the central FOV, we efficiently solved the following minimization problem with Tikhonov regularization added to the deconvolution to suppress noise amplification:49
where * represents convolution, \(\hat{{{{{{\boldsymbol{i}}}}}}}\) is the estimated scene, i is the actual scene, b is the measured signal from the sensor, \(p\) is the PSF at the depth of the scene, \(\gamma\) is the weight of regularization, and ‖·‖F is the Frobenius norm. To minimize computational complexity, this optimization problem can be solved in closed form using Wiener deconvolution as:
where ⨀ represents the Hadamard product, \({(\cdot )}^{*}\) is the complex conjugate operator, \(F\) represents the Fourier transform, and \({F}^{-1}\) denotes the inverse Fourier transform. The processing time on an 8-Core Processor (AMD Ryzen 7 3700X, 3.59 GHz) is approximately 0.5 s for one 8-bit frame with 2048 × 2048 pixels using MATLAB. The fast reconstruction speed allows for real-time feedback on the device position and initial image quality check, making it practical for on-site use.
After the imaging experiment, all captured data were reconstructed using spatially variant PSFs obtained at the imaging depth, resulting in improved reconstruction quality compared to using the shift-invariant model51. The reconstruction depth was selected based on the sharpness of features of interest in the region of interest, with the goal of obtaining the best reconstructed results. The scene i and the background g were jointly estimated by solving a regularized minimization problem with Tikhonov regularization in the deconvolution:
where \(\varPhi\) is the matrix representation of the spatially variant PSFs. The scene i and the low-frequency background g were jointly solved by using FISTA88, with the constraint that the Direct Cosine Transform (DCT) coefficients of g are set to zero outsize the 5 × 5 lowest frequency components. The reconstruction time on a Nvidia GeForce RTX 2070 GPU is approximately 3 min for one 8-bit frame with 2048 × 2048 pixels.
Sample preparation for spiking HEK cells
Spiking HEK 239 cells64 were cultured in DMEM-F12 (Lonza) supplemented with 10% phosphate-buffered saline (Gibso) and 1% penicillin/streptomycin (Lonza) at 37 °C in a 5% CO2 environment. The glass coverslips, with a diameter of 12 mm, were treated with polydimethylsiloxane to create circles 300 μm in diameter, with a 300 μm spacing between them, using a Lumen 3D printing system65,89. The cells were seeded on the coverslips, with 10,000 to 20,000 cells per coverslip, 24−48 h before imaging, resulting in isolated colonies of HEK 293 cells with well-defined size and geometry. The cells were incubated with 2 μM Calcein-AM 30 min prior to imaging. The coverslip was then transferred to a petri dish containing 1 mL PBS for live cell imaging. Both Bio-FlatScopeNHP imaging and ground truth imaging were conducted within 30 min of transferring the coverslip to the petri dish.
Animal for in vivo imaging experiment
All Experiments were approved by University of Texas Institutional Animal Care and Use Committee (IACUC) under protocol lAUP-2023-00063 and conform to NIH standards. One male rhesus monkey (macaca mulatta, 8 years old) was used in this study. The monkey weighted between 9 and 10 kg.
Surgical procedure
All procedures have been approved by the University of Texas Institutional Animal Care and Use Committee and conform to NIH standards. Our general experimental procedures in behaving macaque monkeys have been described in detail previously12,13,47. Briefly, the animal was implanted with a metal head post and a metal recording chamber located over the dorsal portion of V1, a region representing the lower contra-lateral visual field at eccentricities of 2–5 deg. Craniotomy and durotomy were performed in order to obtain a chronic cranial window. A transparent artificial dura made of silicone was used to protect the brain while allowing optical access for imaging.
Details of the method for injecting virus in the animal have been described previously12,13,47. The virtual we used is AAV1-CaMKIIa-NES-GCaMP6f with a titer of 4.8E12 viral genomes per microliter, which only infected the excitatory cells. In summary, a glass pipette (20 μm tip diameter) was first lowered through an opening in the imaging chamber, puncturing the pia. Injections were made at depths of 1.5, 1.0, and 0.5 mm using a Nanoject II. At each depth, 0.5 μL was delivered manually in 10 × 50 nL steps, with approximately 30 s pauses between steps. The injection was performed in a hexagon array with a neighbor distance of 2 mm across an area of 50 ~ 100 mm2, resulting in a consistent and uniform expression. In our previous study13, we showed that the viral expression was relatively uniform within the imaging ROI using this injection strategy. The effect of slight nonuniformity expression will only present in a low spatial frequency, and can be filtered out during the signal processing using a bandpass spatial filter (0.8 cyc/mm cutoff). The quantitative measurements and computational model from our previous study13 suggest that the measured signals reflect the pooled spiking activity of layer 2/3 neurons rather than their pooled synaptic potentials or pre-synaptic inputs.
Recording sessions for in vivo imaging of behaving animals
The experimental techniques for optical imaging in behaving monkeys have been described in detail elsewhere12,13,90,91. We imaged GCaMP signals from V1 of one monkey with a well-shape insert with 8 mm height, whose bottom is 200 μm-thick glass with 16 mm diameter. The insert has an inner diam of 21 mm and an outer diam of 23 mm. The imaging chamber has an outer diameter of 36 mm and an inner diameter of the opening 26 mm. To limit the imaging FOV, a square aperture was constructed from double coated carbon conductive tape (TED PELLA, INC) and placed at the bottom of the chamber insert. For position tuning imaging, the aperture size was 5 mm × 5 mm, and for orientation imaging the aperture size was 4.5 mm × 4.5 mm. The ground truth epi-fluorescence imaging was performed with a custom designed imaging system based on an sCMOS camera (PCO4.2 camera) using the following filter sets: GCaMP, excitation 470/24 nm, dichroic 505 nm, emission 515 nm cutoff glass filter. Illumination was obtained with an LED light source (X-Cite 110 LED). The Bio-FlatScopeNHP imaging was performed in the same recording session, and illumination was obtained by using the integrated light source on the device. The Bio-FlatScopeNHP was mounted on top of the implant by using two 8–32 screws embedded in dental acrylic (Supplementary Fig 5c, d). Data acquisition was time locked to the animal’s heartbeat. Slow hemodynamic signals usually start 2 s after stimulus onset and peaks at 4 ~ 6 s4,6,92, which is much longer than the stimulus and imaging duration used in this study. Within the short stimulus time, the largest artifact is the heartbeats (at 2–3 Hz)93, and this artifact can be reduced by synchronizing the data acquisition to the electrocardiogram (Supplementary Fig. 15). This allowed us to remove the average blank time course from all trials, which significantly reduced the fast hemodynamic artifacts. The imaging was performed under the same conditions and within a two-hour time window. Both devices were recorded at 20 Hz for GCaMP imaging. For both position and orientation tuning experiments, visual stimulus conditions were randomly interleaved, repeated 10 times each, and mixed with blank fixation trials.
Imaging stability on head-unrestrained animals
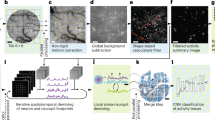
We have evaluated the stability of the images captured using the Bio-FlatScopeNHP during head-unrestrained recording. We captured an image before releasing the head restriction of the animal, and this image was used as the template for motion correction. We analyzed the motion in 10 trials in one head-unrestrained experiment session. The absolute maximum displacements in the medial-lateral and anterior-posterior direction were 17.2 μm and 27.2 μm, respectively (Supplementary Fig 13). These small displacements caused by motion could be digitally corrected by registering each frame using vascular structures.
Data analysis for imaging position tuning
The process for analyzing the ground truth signals captured by the widefield microscope involved several steps. First, the average time course in blank trials was subtracted from the response in each stimulus condition. Next, the mean residual response in the 200 ms period before response onset was removed to reduce the effect of sources of noise like the heartbeat artifact and other slow, widespread fluctuations in the signals. The imaging signals were then averaged across repeats. To compute the spatial tuning curves, six regions of interest (ROIs) were selected, each with a size of 0.3 × 0.3 mm2 and spaced approximately 0.37 mm apart (as shown in Fig. 3d, e). The average response from the ROIs was fitted with a 1D Gaussian function. The Bio-FlatScopeNHP images were reconstructed frame by frame from the raw captured videos using spatially variant PSFs, and downsized to the same size as images captured by the ground truth microscope. The analysis process was identical to that carried out on the widefield microscope images. The data analysis was conducted using Matlab.
Data analysis for imaging orientation tuning
The Bio-FlatScopeNHP images were obtained by reconstructing each frame of the raw captured videos using spatially variant PSFs, followed by downsizing them to the same size as images captured by the ground truth microscope. The remaining data analysis process was identical for both types of images, and was conducted using Matlab. Initially, the blank trials were subtracted, and the mean residual response was removed, after which the data was averaged, and this processing was the same as that used for processing position tuning data. The ROI was selected based on the RMS map of the response, and the areas with the strongest 4 Hz signals were identified by choosing the overlap** regions between the ground truth and Bio-FlatScopeNHP captures, where the RMS was greater than one-third of the maximum value (Supplementary Fig 9). Once the ROIs were selected, we obtained the orientation response by calculating the first harmonic amplitude using Fast Fourier Transform (FFT) of the temporal response. To study the spatial scale of the orientation columns, we selected a range of spatial frequencies based on the average 1D amplitude spectrum66 (Figs. 4f, 5d). The peak at approximately 1.2 cycles/mm corresponds to the periodicity of the orientation columns, with an average cycle of around 0.83 mm. To remove any non-orientation-selective responses and high frequency noise, we applied a bandpass spatial filter of 0.8–2.5 cycle/mm. This spatial filtration removes components with periods larger than 1.25 mm and smaller than 0.4 mm.
The mean and standard deviation of the response image at each pixel location were then calculated separately for each condition in each session94. The d’ map was calculated by using the mean-response image of 0 degree \({m}_{0}(x,y)\) and 90 degree \({m}_{90}(x,y)\) and SD-response image of 0 degree \({\sigma }_{0}(x,y)\) and 90 degree \({\sigma }_{90}(x,y)\):
The decision variables DV (Fig. 4e) were then calculated by using the weights equal to the d’ maps:
The discriminability between 0 degree and 90 degree can be calculated as:
where \({m}_{{DV}0}\) and \({m}_{{DV}90}\) are the average of 0 degree and 90 degree decision variables, \({\sigma }_{{DV}0}\) and \({\sigma }_{{DV}90}\) are the standard deviations of 0 degree and 90 degree decision variables.
To verify that the resulting maps reflected the orientation columns, we calculated the correlation between pairs of maps as a function of their stimulus orientation differences (Figs. 4g & 5e). Next, we used a vector summation method66 to obtain the complete orientation map from single orientation response maps (Figs. 4h & 5f). In these maps, color represents the preferred orientation at each location, and the saturation indicates the strength of orientation tuning.
Statistics and Reproducibility
For Fig. 2c, d, the imaging of the USAF test target and Convallaria with both the Bio-FlatScopeNHP and 4x objective were repeated more than 10 times, with similar results. For Fig. 2e, f, the imaging of patterned live spiking HEK293 cells was performed 3 times, with similar results. For Fig. 3f, the position tuning experiments were performed 2 times on 2 different days on a head-fixed male macaque, with similar results. For Fig. 4c, h, the orientation tuning experiments were performed 2 times on 2 different days on a head-fixed male macaque, with similar results. For Fig. 5c, f, the orientation tuning experiments were performed 2 times on the same day on a head-unrestrained male macaque, with similar results. Additional results were presented in Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The main data supporting the results of this study are available within the paper and its Supplementary information. Bio-FlatScopeNHP CAD design, phase mask design, all costume codes and sample data are provided on a Github repository95. The raw and analyzed datasets generated during the study are too large to be publicly shared, but they are available for research purposes from the corresponding author upon request. Source data are provided with this paper.
Code availability
Custom MATLAB codes are available on a GitHub repository95 (https://github.com/JiminWu/Bio-FlatScopeNHP).
References
Urai, A. E., Doiron, B., Leifer, A. M. & Churchland, A. K. Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 25, 11–19 (2022).
Cardin, J. A., Crair, M. C. & Higley, M. J. Mesoscopic Imaging: Shining a Wide Light on Large-Scale Neural Dynamics. Neuron 108, 33–43 (2020).
Lake, E. M. R. et al. Simultaneous cortex-wide fluorescence Ca2+ imaging and whole-brain fMRI. Nat. Methods 17, 1262–1271 (2020).
Ma, Y. et al. Wide-field optical map** of neural activity and brain haemodynamics: considerations and novel approaches. Philos. Trans. R. Soc. B Biol. Sci. 371, 20150360 (2016).
Das, A. & Gilbert, C. D. Long-range horizontal connections and their role in cortical reorganization revealed by optical recording of cat primary visual cortex. Nature 375, 780–784 (1995).
Grinvald, A., Lieke, E., Frostig, R. D., Gilbert, C. D. & Wiesel, T. N. Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature 324, 361–364 (1986).
Bonhoeffer, T. & Grinvald, A. lso-orientation domains in cat visual cortex are arranged in pinwheel-like patterns. Nature 353, 429–431 (1991).
Arieli, A., Sterkin, A., Grinvald, A. & Aertsen, A. Dynamics of Ongoing Activity: Explanation of the Large Variability in Evoked Cortical Responses. Science 273, 1868–1871 (1996).
Weliky, M., Bosking, W. H. & Fitzpatrick, D. A systematic map of direction preference in primary visual cortex. Nature 379, 725–728 (1996).
Ts’o, D. Y., Frostig, R. D., Lieke, E. E. & Grinvald, A. Functional Organization of Primate Visual Cortex Revealed by High Resolution Optical Imaging. Science 249, 417–420 (1990).
Seidemann, E., Arieli, A., Grinvald, A. & Slovin, H. Dynamics of Depolarization and Hyperpolarization in the Frontal Cortex and Saccade Goal. Science 295, 862–865 (2002).
Chen, Y., Geisler, W. S. & Seidemann, E. Optimal decoding of correlated neural population responses in the primate visual cortex. Nat. Neurosci. 9, 1412–1420 (2006).
Seidemann, E. et al. Calcium imaging with genetically encoded indicators in behaving primates. eLife 5, e16178 (2016).
Vnek, N., Ramsden, B. M., Hung, C. P., Goldman-Rakic, P. S. & Roe, A. W. Optical imaging of functional domains in the cortex of the awake and behaving monkey. Proc. Natl Acad. Sci. 96, 4057–4060 (1999).
Blasdel, G. G. & Salama, G. Voltage-sensitive dyes reveal a modular organization in monkey striate cortex. Nature 321, 579–585 (1986).
Capitanio, J. P. & Emborg, M. E. Contributions of non-human primates to neuroscience research. Lancet 371, 1126–1135 (2008).
Phillips, K. A. et al. Why primate models matter: Why Primate Models Matter. Am. J. Primatol. 76, 801–827 (2014).
Roelfsema, P. R. & Treue, S. Basic Neuroscience Research with Nonhuman Primates: A Small but Indispensable Component of Biomedical Research. Neuron 82, 1200–1204 (2014).
Jennings, J. H. & Stuber, G. D. Tools for Resolving Functional Activity and Connectivity within Intact Neural Circuits. Curr. Biol. 24, R41–R50 (2014).
Hamel, E. J. O., Grewe, B. F., Parker, J. G. & Schnitzer, M. J. Cellular Level Brain Imaging in Behaving Mammals: An Engineering Approach. Neuron 86, 140–159 (2015).
Yang, W. & Yuste, R. In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017).
Hubel, D. H. & Wiesel, T. N. Receptive fields and functional architecture of monkey striate cortex. J. Physiol. 195, 215–243 (1968).
Obermayer, K. & Blasdel, G. Geometry of orientation and ocular dominance columns in monkey striate cortex. J. Neurosci. 13, 4114–4129 (1993).
Ohki, K., Chung, S., Ch’ng, Y. H., Kara, P. & Reid, R. C. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603 (2005).
Palmer, C. R., Chen, Y. & Seidemann, E. Uniform spatial spread of population activity in primate parafoveal V1. J. Neurophysiol. 107, 1857–1867 (2012).
Van Hooser, S. D., Heimel, J. A. F., Chung, S., Nelson, S. B. & Toth, L. J. Orientation Selectivity without Orientation Maps in Visual Cortex of a Highly Visual Mammal. J. Neurosci. 25, 19–28 (2005).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Luo, L., Callaway, E. M. & Svoboda, K. Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 256–281 (2018).
Sadakane, O. et al. Long-Term Two-Photon Calcium Imaging of Neuronal Populations with Subcellular Resolution in Adult Non-human Primates. Cell Rep. 13, 1989–1999 (2015).
Yamada, Y., Matsumoto, Y., Okahara, N. & Mikoshiba, K. Chronic multiscale imaging of neuronal activity in the awake common marmoset. Sci. Rep. 6, 35722 (2016).
Santisakultarm, T. P. et al. Two-photon imaging of cerebral hemodynamics and neural activity in awake and anesthetized marmosets. J. Neurosci. Methods 271, 55–64 (2016).
Ebina, T. et al. Two-photon imaging of neuronal activity in motor cortex of marmosets during upper-limb movement tasks. Nat. Commun. 9, 1879 (2018).
Li, M., Liu, F., Jiang, H., Lee, T. S. & Tang, S. Long-Term Two-Photon Imaging in Awake Macaque Monkey. Neuron 93, 1049–1057.e3 (2017).
Trautmann, E. M. et al. Dendritic calcium signals in rhesus macaque motor cortex drive an optical brain-computer interface. Nat. Commun. 12, 3689 (2021).
**e, Y. et al. Geometry of sequence working memory in macaque prefrontal cortex. Science 375, 632–639 (2022).
Meyer, A. F., O’Keefe, J. & Poort, J. Two Distinct Types of Eye-Head Coupling in Freely Moving Mice. Curr. Biol. 30, 2116–2130.e6 (2020).
Zong, W. et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017).
Skocek, O. et al. High-speed volumetric imaging of neuronal activity in freely moving rodents. Nat. Methods 15, 429–432 (2018).
Ziv, Y. et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 16, 264–266 (2013).
Ghosh, K. K. et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011).
Rynes, M. L. et al. Miniaturized head-mounted microscope for whole-cortex mesoscale imaging in freely behaving mice. Nat. Methods 18, 417–425 (2021).
Kondo, T. et al. Calcium Transient Dynamics of Neural Ensembles in the Primary Motor Cortex of Naturally Behaving Monkeys. Cell Rep. 24, 2191–2195.e4 (2018).
Bollimunta, A. et al. Head-mounted microendoscopic calcium imaging in dorsal premotor cortex of behaving rhesus macaque. Cell Rep. 35, 109239 (2021).
Hubel, D. H. & Wiesel, T. N. Uniformity of monkey striate cortex: A parallel relationship between field size, scatter, and magnification factor. J. Comp. Neurol. 158, 295–305 (1974).
Seidemann, E. & Geisler, W. S. Linking V1 Activity to Behavior. Annu. Rev. Vis. Sci. 4, 287–310 (2018).
Van Essen, D. C., Newsome, W. T. & Maunsell, J. H. R. The visual field representation in striate cortex of the macaque monkey: Asymmetries, anisotropies, and individual variability. Vis. Res. 24, 429–448 (1984).
Chen, Y., Geisler, W. S. & Seidemann, E. Optimal Temporal Decoding of Neural Population Responses in a Reaction-Time Visual Detection Task. J. Neurophysiol. 99, 1366–1379 (2008).
Zaraza, D. et al. Head-mounted optical imaging and optogenetic stimulation system for use in behaving primates. Cell Rep. Methods 2, 100351 (2022).
Adams, J. K. et al. In vivo lensless microscopy via a phase mask generating diffraction patterns with high-contrast contours. Nat. Biomed. Eng. 6, 617–628 (2022).
Adams, J. K. et al. Single-frame 3D fluorescence microscopy with ultraminiature lensless FlatScope. Sci. Adv. 3, e1701548 (2017).
Kuo, G., Linda Liu, F., Grossrubatscher, I., Ng, R. & Waller, L. On-chip fluorescence microscopy with a random microlens diffuser. Opt. Express 28, 8384 (2020).
Boominathan, V., Robinson, J. T., Waller, L. & Veeraraghavan, A. Recent advances in lensless imaging. Optica 9, 1 (2022).
Antipa, N. et al. DiffuserCam: lensless single-exposure 3D imaging. Optica 5, 1 (2018).
Xue, Y., Davison, I. G., Boas, D. A. & Tian, L. Single-shot 3D wide-field fluorescence imaging with a Computational Miniature Mesoscope. Sci. Adv. 6, eabb7508 (2020).
Xue, Y., Yang, Q., Hu, G., Guo, K. & Tian, L. Deep-learning-augmented computational miniature mesoscope. Optica 9, 1009 (2022).
Wu, J., Boominathan, V., Veeraraghavan, A. & Robinson, J. T. Real-time, deep-learning aided lensless microscope. Biomed. Opt. Express 14, 4037 (2023).
Yanny, K. et al. Miniscope3D: optimized single-shot miniature 3D fluorescence microscopy. Light Sci. Appl. 9, 171 (2020).
Boominathan, V., Adams, J. K., Robinson, J. T. & Veeraraghavan, A. PhlatCam: Designed Phase-Mask Based Thin Lensless Camera. IEEE Trans. Pattern Anal. Mach. Intell. 42, 1618–1629 (2020).
Scott, B. B. et al. Imaging Cortical Dynamics in GCaMP Transgenic Rats with a Head-Mounted Widefield Macroscope. Neuron 100, 1045–1058.e5 (2018).
Zhou, Z. C., Yu, C., Sellers, K. K. & Fröhlich, F. Dorso-Lateral Frontal Cortex of the Ferret Encodes Perceptual Difficulty during Visual Discrimination. Sci. Rep. 6, 23568 (2016).
Walker, J. D. et al. Chronic wireless neural population recordings with common marmosets. Cell Rep. 36, 109379 (2021).
Richard, C., Renaudin, A., Aimez, V. & Charette, P. G. An integrated hybrid interference and absorption filter for fluorescence detection in lab-on-a-chip devices. Lab. Chip 9, 1371 (2009).
Sasagawa, K. et al. Highly sensitive lens-free fluorescence imaging device enabled by a complementary combination of interference and absorption filters. Biomed. Opt. Express 9, 4329 (2018).
Park, J. et al. Screening Fluorescent Voltage Indicators with Spontaneously Spiking HEK Cells. PLoS ONE 8, e85221 (2013).
Zhu, Y., Sazer, D., Miller, J. S. & Warmflash, A. Rapid fabrication of hydrogel micropatterns by projection stereolithography for studying self-organized developmental patterning. PLOS ONE 16, e0245634 (2021).
Chen, Y., Palmer, C. R. & Seidemann, E. The relationship between voltage-sensitive dye imaging signals and spiking activity of neural populations in primate V1. J. Neurophysiol. 107, 3281–3295 (2012).
Adams, D. L. & Horton, J. C. A Precise Retinotopic Map of Primate Striate Cortex Generated from the Representation of Angioscotomas. J. Neurosci. 23, 3771–3789 (2003).
Yang, Z., Heeger, D. J. & Seidemann, E. Rapid and Precise Retinotopic Map** of the Visual Cortex Obtained by Voltage-Sensitive Dye Imaging in the Behaving Monkey. J. Neurophysiol. 98, 1002–1014 (2007).
Jung, Y. J. et al. Orientation pinwheels in primary visual cortex of a highly visual marsupial. Sci. Adv. 8, eabn0954 (2022).
Jay, M. F. & Sparks, D. L. Auditory receptive fields in primate superior colliculus shift with changes in eye position. Nature 309, 345–347 (1984).
Klier, E. M. & Crawford, J. D. Neural Control of Three-Dimensional Eye and Head Posture. Ann. N. Y. Acad. Sci. 1004, 122–131 (2003).
Churchland, M. M. et al. Neural population dynamics during reaching. Nature 487, 51–56 (2012).
Buneo, C. A., Jarvis, M. R., Batista, A. P. & Andersen, R. A. Direct visuomotor transformations for reaching. Nature 416, 632–636 (2002).
Báez-Mendoza, R., Mastrobattista, E. P., Wang, A. J. & Williams, Z. M. Social agent identity cells in the prefrontal cortex of interacting groups of primates. Science 374, eabb4149 (2021).
Sliwa, J. & Freiwald, W. A. A dedicated network for social interaction processing in the primate brain. Science 356, 745–749 (2017).
Barros, M. & Tomaz, C. Non-human primate models for investigating fear and anxiety. Neurosci. Biobehav. Rev. 26, 187–201 (2002).
Shuman, T. et al. Breakdown of spatial coding and interneuron synchronization in epileptic mice. Nat. Neurosci. 23, 229–238 (2020).
Guo, C. et al. Miniscope-LFOV: A large-field-of-view, single-cell-resolution, miniature microscope for wired and wire-free imaging of neural dynamics in freely behaving animals. Sci. Adv. 9, eadg3918 (2023).
Leman, D. P. et al. Large-scale cellular-resolution imaging of neural activity in freely behaving mice. https://doi.org/10.1101/2021.01.15.426462 (2021).
Scherrer, J. R., Lynch, G. F., Zhang, J. J. & Fee, M. S. An optical design enabling lightweight and large field-of-view head-mounted microscopes. Nat. Methods 20, 546–549 (2023).
Kim, T. H. & Schnitzer, M. J. Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185, 9–41 (2022).
Mehta, P. et al. Functional Access to Neuron Subclasses in Rodent and Primate Forebrain. Cell Rep. 26, 2818–2832.e8 (2019).
Vormstein-Schneider, D. et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci. 23, 1629–1636 (2020).
Cushnie, A. K. et al. Using rAAV2-retro in rhesus macaques: Promise and caveats for circuit manipulation. J. Neurosci. Methods 345, 108859 (2020).
Weiss, A. R., Liguore, W. A., Domire, J. S., Button, D. & McBride, J. L. Intra-striatal AAV2.retro administration leads to extensive retrograde transport in the rhesus macaque brain: implications for disease modeling and therapeutic development. Sci. Rep. 10, 6970 (2020).
Pollmann, E. H. et al. Subdural CMOS optical probe (SCOPe) for bidirectional neural interfacing. https://doi.org/10.1101/2023.02.07.527500 (2023).
Kwon, Seonil et al. Development of high performance, flexible, and transparent, bio-implantable OLEDs for in vivo optogenetics. (2022).
Beck, A. & Teboulle, M. A Fast Iterative Shrinkage-Thresholding Algorithm for Linear Inverse Problems. SIAM J. Imaging Sci. 2, 183–202 (2009).
Grigoryan, B. et al. Development, characterization, and applications of multi-material stereolithography bioprinting. Sci. Rep. 11, 3171 (2021).
Arieli, A., Grinvald, A. & Slovin, H. Dural substitute for long-term imaging of cortical activity in behaving monkeys and its clinical implications. J. Neurosci. Methods 114, 119–133 (2002).
Shtoyerman, E., Arieli, A., Slovin, H., Vanzetta, I. & Grinvald, A. Long-Term Optical Imaging and Spectroscopy Reveal Mechanisms Underlying the Intrinsic Signal and Stability of Cortical Maps in V1 of Behaving Monkeys. J. Neurosci. 20, 8111–8121 (2000).
Frostig, R. In Vivo Optical Imaging of Brain Function. (CRC Press, 2009).
Grinvald, A. et al. In-vivo Optical Imaging of Cortical Architecture and Dynamics. in Modern Techniques in Neuroscience Research (eds. Windhorst, U. & Johansson, H.) 893–969 (Springer Berlin Heidelberg, 1999). https://doi.org/10.1007/978-3-642-58552-4_34.
Benvenuti, G. et al. Scale-Invariant Visual Capabilities Explained by Topographic Representations of Luminance and Texture in Primate V1. Neuron 100, 1504–1512.e4 (2018).
Wu, J., Chen, Y., Veeraraghavan, A., Seidemann, E. & Robinson, J. T. Mesoscopic calcium imaging in a head-unrestrained non-human male primate using a lensless microscope, Bio-FlatScopeNHP, https://doi.org/10.5281/zenodo.10403840 (2023).
Acknowledgements
This work was supported in part by DARPA grant N66001-17-C-4012 for JTR AV & ES, NIH grant RF1NS110501 for J.T.R. & A.V., NSF grant IIS-1730574 and IIS-1652633 for A.V., NIH grants R01EY016454 and U19NS118284 for E.S.; This research is partially sponsored by the Defense Advanced Research Projects Agency (DARPA) through Cooperative Agreement D20AC00002 (for J.T.R. & A.V.) awarded by the U.S. Department of the Interior (DOI), Interior Business Center. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. We thank G. Duret for preparing the Spiking HEK cells samples; V. Boominathan for helpful discussions and the scanned electron micrograph of the phase mask; and D. Yan for the help on calibration setups.
Author information
Authors and Affiliations
Contributions
J.W. designed, fabricated, and characterized the Bio-FlatScopeNHP prototype, and developed the reconstruction model. Y.C. designed and constructed the table-top wide-field imaging system, performed virus injections, and performed animal training. Y.C. and E.S. designed the in vivo experiments with visual stimulus. J.W. and Y.C. performed the in vivo imaging experiments and data analysis. A.V., E.S., and J.T.R. provided guidance and assistance with all aspects of the work. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.T.R. is cofounder of, holds equity in, and receives payment from Motif Neurotech. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Aniruddha Das and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, J., Chen, Y., Veeraraghavan, A. et al. Mesoscopic calcium imaging in a head-unrestrained male non-human primate using a lensless microscope. Nat Commun 15, 1271 (2024). https://doi.org/10.1038/s41467-024-45417-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-45417-6
- Springer Nature Limited