Abstract
In response to DNA double-strand breaks (DSBs), DNA damage repair factors are recruited to DNA lesions and form nuclear foci. However, the underlying molecular mechanism remains largely elusive. Here, by analyzing the localization of DSB repair factors in the XY body and DSB foci, we demonstrate that pre-ribosomal RNA (pre-rRNA) mediates the recruitment of DSB repair factors around DNA lesions. Pre-rRNA exists in the XY body, a DSB repair hub, during meiotic prophase, and colocalizes with DSB repair factors, such as MDC1, BRCA1 and TopBP1. Moreover, pre-rRNA-associated proteins and RNAs, such as ribosomal protein subunits, RNase MRP and snoRNAs, also localize in the XY body. Similar to those in the XY body, pre-rRNA and ribosomal proteins also localize at DSB foci and associate with DSB repair factors. RNA polymerase I inhibitor treatment that transiently suppresses transcription of rDNA but does not affect global protein translation abolishes foci formation of DSB repair factors as well as DSB repair. The FHA domain and PST repeats of MDC1 recognize pre-rRNA and mediate phase separation of DSB repair factors, which may be the molecular basis for the foci formation of DSB repair factors during DSB response.
Similar content being viewed by others
Introduction
Once DNA double-strand breaks (DSBs) occur, DNA repair factors are recruited to DNA lesions and repair DSBs via different pathways.1,2 These DSB repair factors are sequentially loaded onto DNA lesions and form distinct condensates in nucleus, known as DNA damage-induced foci.3 This phenomenon is often observed and examined when cells are treated with ionizing radiation to induce DSBs.4 Thus, it is also known as ionizing radiation-induced foci (IRIF).
The foci formation of DSB repair factors is maintained by phosphorylation of H2AX, a variant of histone H2A, at Ser139 (aka γH2AX).5,6 In response to DSBs, this site-specific phosphorylation event is mediated by a group of PI3-like kinases, including ATM, ATR and DNA-PK.3,7 γH2AX may extend to mega-base away from DNA lesions, and provides a platform to cluster DSB repair factors at vicinity of DSBs.8,9 Although the detailed molecular mechanism is unclear, loss of H2AX abolishes DNA damage-induced foci and impairs DSB repair.10,11,12
In addition to environmental hazard-induced DSBs, DSBs also arise during physiologically relevant processes. A typical example is homologous recombination (HR) before meiosis. In both autosomes and sex chromosomes of germ cells, DSBs are generated by SPO11, a topoisomerase-like enzyme.13,14,15,16,17 However, unlike topoisomerases, SPO11 cannot religate DSBs. Instead, these DSBs are repaired by DSB repair factors to complete HR. Due to the lack of homology, the X and Y chromosomes in male germ cells cannot use canonical HR to repair SPO11-induced DSBs.18 Thus, DSBs on the X and Y chromosomes exist for prolonged time, which causes extensive DNA damage response. Various DNA damage repair factors are concentrated into these two sex chromosomes to form a phase separation-like structure, named the XY body, during meiotic prophase.18
This phase separation-like structure covers the whole X and Y chromosomes. As the obvious biomarker of the XY body is γH2AX, we have proposed that the XY body is by far the largest DNA damage focus in human cells.18,37 and we did not detect obvious staining of the XY body using this antibody (Supplementary information, Fig. S3a). Since RNase H specifically digests RNA that forms R-loop, we also treated meiotic spreads with RNase H, and found that RNase H did not affect the RNA staining in the XY body (Supplementary information, Fig. S3b). Thus, these results collectively suggest that the RNA species in the XY body may not form DNA-RNA hybrid.
RNA species observed in the XY body are more abundant than those in other part of nucleus, indicating that these RNA species should be some of the most abundant RNAs in the cell. Since ribosomal RNA (rRNA) is the most abundant RNA, we examined rRNA localization in meiotic spreads with anti-rRNA antibody. This monoclonal antibody specifically recognizes 5.8S rRNA. Interestingly, we found that rRNA was remarkably enriched in the XY body (Fig. 1d). Since it could not be digested by RNase H, the rRNA in XY body did not form R-loops (Supplementary information, Fig. S3c). Instead, with the RNase A treatment, the rRNA staining was disappeared from meiotic spreads. Using excessive rRNA to block the epitope recognized by this antibody, the rRNA in the XY body could not be stained (Fig. 1e). Since γH2AX mainly exists in the XY body, we harvested spermatogonia from 3-week-old male mice, and performed RNA sequencing (RNA-seq) following RNA-chromatin immunoprecipitation assay (RNA-ChIP) using anti-γH2AX antibody (Supplementary information, Fig. S4a). We found that majorities RNA associated with γH2AX was rRNA (Supplementary information, Fig. S4b, c). These results were further validated by real-time quantitative PCR (RT-qPCR) (Supplementary information, Fig. S4d). Collectively, these results suggest that rRNA exists in the XY body.
Pre-rRNAs exist in the XY body
rRNA is transcribed from ribosomal DNA (rDNA) loci as 47S pre-rRNA by RNA Polymerase I (RNA pol I). Immediately following transcription, 47S pre-rRNA is processed to 45S pre-rRNA that is further digested into 32S and 34S pre-rRNA in murine cells, the precursors of 18S, 5.8S and 28S rRNA.38 Once pre-rRNA is transcribed, it undergoes extensive modifications such as 2’-O-methylation and pseudouridylation.39 Moreover, protein partners and 5S rRNA, the smallest rRNA component transcribed by RNA Polymerase III (RNA pol III), are incorporated into pre-rRNA to form pre-ribonucleoprotein particles (pre-rRNPs).40,41 To examine the rRNA species in the XY body, we designed various probes covering different regions of 45S and 5S rRNA (Fig. 2a). Interestingly, we could stain rRNA with probes targeting the regions of 34S, 18S, 18SE, 32S, 28S, 12S, 8S, 5.8S and 5S but not with 5’-ETS probes (Fig. 2b), suggesting that the XY body contains 34S, 32S rRNA and probably their derivatives, but not 45S rRNA. We also hybridized different probes simultaneously and found that these rRNAs had the same signal distribution pattern in the XY body (Supplementary information, Fig. S5a–c). To examine the component of rRNA species in the XY body, we measured the fluorescence intensity of each probe in the XY body (Supplementary information, Fig. S5d). Based on the relative signal intensity, we calculated and found that these rRNA species mainly included 34S (18.32%), 32S (80.27%) and 5S (1.41%) rRNAs (Fig. 2c). Although probes targeting other regions also recognized the XY body, the staining of other probes mainly reflected 34S and 32S pre-rRNAs.
a The sketches of pre-rRNA processing and location of each RNA probe. Different probe recognizes different pre-rRNAs or mature rRNA. For example, probe 1 only recognizes 45S pre-rRNA, while probe 2 can recognize 18S, 18SE, 34S and 45S. b Localization of rRNA in prophase spermatocytes was examined by indicated rRNA anti-sense probes in the in situ hybridization. c rRNA species in the XY body were calculated. The calculation was based on the relative fluorescence intensity and molecular weight of each species. d Different patterns of pre-rRNA distribution in meiotic prophase. e Pre-rRNA distribution from early pachytene to diplotene is statistically analyzed. f Pre-rRNA is enriched in the unsynapsed axes of X and Y chromosomes. g Analysis of rRNA and SCP3 distribution of f. The initial point 0 indicated the center of unsynapsed axes of X and Y chromosomes. Both rRNA and SCP3 are enriched in the unsynapsed axes. Circled area indicates the XY body. Scale bars, 10 μm.
To exclude any mature rRNA in the XY body, we used probe quenchers that could interact with adjacent 5.8S or 28S probe, which sufficiently abolished the fluorescence signals of 5.8S or 28S probe, suggesting that most of the 5.8S or 28S probes recognize 5.8S or 28S region in pre-rRNAs. Thus, the results indicate that pre-rRNAs are dominant RNA species in the XY body and little mature rRNA exists in the XY body (Supplementary information, Fig. S5e).
In the XY body, pre-rRNA localized into different patterns, and we summarized these localization patterns into four types: (1) pre-rRNA localizes at the edge of the XY body; (2) pre-rRNA forms a cap on the XY body; (3) pre-rRNA partially occupies the XY body; (4) pre-rRNA exists in the whole XY body (Fig. 2d). Interestingly, we found that the localization pattern of rRNA was shifted from the edge of the XY body in early pachytene and gradually migrated into the XY body till diplotene (Fig. 2e). Moreover, when pre-rRNA entered the XY body, it was enriched at the unsynapsed axes of the X and Y chromosomes (Fig. 2f, g). However, as conventional paraformaldehyde fixation crosslinks proteins and nucleic acids in the XY body, it may impair RNA probe hybridization. We also used methanol/acetic acid to fix cells, in which acetic acid treatment removes proteins and exposes nucleic acids during fixation. we found that pre-RNA species formed a string pattern and might be enriched at the unsynapsed axes of the X and Y chromosomes (Supplementary information, Fig. S5f). In addition, we also examined other stages of meiotic prophase before the XY body formation, including leptotene and zygotene. However, we did not observe obvious rRNA staining in these stages (Supplementary information, Fig. S6), suggesting that pre-rRNA is highly enriched in the XY body during pachytene and diplotene.
Pre-rRNA-associated RNA and proteins localize in the XY body
Once pre-rRNA is transcribed, it associates with other small nucleolar RNAs (snoRNA) and nucleolar proteins to form pre-rRNPs.42 Thus, we asked whether snoRNAs- and pre-rRNA-associated proteins exist in the XY body. We found that snoRNA U17, an H/ACA box snoRNA for peudouridylation of pre-rRNA, localized in the XY body (Fig. 3a). In addition, RNase MRP (aka RMRP), a key ribozyme to process pre-rRNA, was also in the XY body (Fig. 3a). Moreover, we examined ribosomal proteins that associate with pre-rRNAs, and found that RPL4, 6, 7A, 8, 13, 14 and RPS3 existed in the XY body (Fig. 3a–d). Among these ribosomal proteins, RPL6, RPL7A and RPS3 clearly localized at the unsynapsed axes of the X and Y chromosomes (Fig. 3b–d). In addition, we found that fibrillarin, rRNA 2’-O-methyltransferase, also sharply localized at the unsynapsed axes of the X and Y chromosomes (Fig. 3e). Other nucleolus proteins including nuclephosmin (NPM1) and nucleolin localized in the XY body too (Supplementary information, Fig. S7). The colocalization of pre-rRNA- and pre-rRNA-associated RNAs/proteins was further confirmed in the XY body (Supplementary information, Fig. S8). Collectively, these results suggest that pre-rRNPs and nucleolus components are associated with the XY body.
a Localization of pre-rRNA-associated RNA and proteins in XY body. SnoRNA U17, RMRP, RPL4, RPL8, RPL13 and RPL14 were examined by RNA probes or indicated antibodies in the late pachytene cells. Percentages of positive staining in the XY body are shown. b–e RPL7A (b), RPL6 (c), RPS3 (d) and FBL (e) are localized on the unsynapsed regions of the X and Y chromosomes. The fluorescence intensity on the white dash line is plotted. Circled area indicates the XY body. Scale bars, 10 μm.
Pre-rRNA associates with DSB repair factors in the XY body
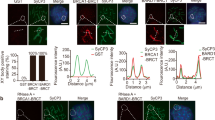
In addition to pre-rRNPs, many DSB repair factors also localize in the XY body. Based on the precise localization in the XY body, these repair factors are classified into two groups. The first group includes γH2AX, MDC1, RNF8, etc., which mediates DNA damage response and exists in the whole XY body. The second group includes BRCA1, TOPBP1, RAD51, etc., which mediates HR repair and only localizes at the unsynapsed axes of the X and Y chromosomes.8b). Moreover, recombinant FHA or PST associated with pre-rRNA to form phase separation droplet in vitro (Fig. 8c–e and Supplementary information, Fig. S15), while RNase A treatment repressed the liquid-liquid phase separation (LLPS) formation (Fig. 8f). Finally, reconstitution of the MDC1-deificent cells with WT MDC1, but not the ΔFHA or ΔPST mutant, could rescue the IRIF of pre-rRNPs (Fig. 8g and Supplementary information, Fig. S16). Taken together, these results suggest that MDC1 interacts with pre-rRNA for phase separation of DSB repair factors at DNA lesions (Supplementary information, Fig. S17).
a MDC1 is associated with pre-rRNA. Anti-MDC1 antibody was used to pull down pre-rRNA in cells with or without 20 Gy of IR. Following anti-MDC1 pull-down assays, qPCR was performed to examine the interaction between MDC1 and pre-rRNA. Primers of ITS1 and ITS2, two regions of pre-rRNA, were applied for qPCR. b The FHA domain and the PDT repeats of MDC1 bind to pre-rRNA. GST, GST-MDC1-FHA, GST-MDC1-PST or GST-MDC1-BRCT proteins were used to pull down pre-rRNA. Following GST pull-down assays, qPCR was performed to examine the enrichment of pre-rRNA. c, d LLPS of the MDC1–pre-rRNA complex. Recombinant MDC1-FHA or MDC1-PST was incubated with pre-rRNA in the presence of a crowding agent (10% PEG 8000). LLPS was examined by microscopy. e Microscopy images of individual droplet collisions at indicated time points. f LLPS of the MDC1–pre-rRNA complex is abolished by RNase A treatment. Recombinant MDC1-FHA and MDC1-PST were incubated with pre-rRNA, followed by 1 mg/mL RNase A treatment. Statistical analysis of LLPS formation is shown in the right panel. g Full-length MDC1 (FL) but not the FHA domain or the PST repeats deletion mutants restores IRIF in MDC1−/− MEFs. MDC1−/− MEFs were reconstituted with FL, ΔPST or ΔFHA mutants. IRIF of pre-rRNA and MDC1 were examined by RNA probes and anti-FLAG antibody. The foci number of pre-rRNA was measured. **P < 0.01, ***P < 0.001. Scale bars, 10 μm.
Discussion
In this work, we have shown that pre-rRNA participates in phase separation of DNA damage repair factors at DNA lesions, taking advantage of the XY body. As mentioned, the XY body serves as a hub to retain various DSB repair factors. Some of these repair factors are covered in the whole area of the XY body, while others localize at the unsynapsed axes of the X and Y chromosomes.19 RAD51 recognizes ssDNA processed from the DSB ends and only localizes at the unsynapsed axes,19 indicating that SPO11-induced DSBs exists at the unsynapsed axes. In contrast, γH2AX is known to extend to mega-base away from DNA lesion,9 possibly accounting for its whole XY body localization. Super-resolution microscopy on IRIF also suggests that γH2AX localizes in the surrounding area, whereas RAD51 exists in the central region of a solo DSB site.22 Interesting, pre-rRNA not only exists in the whole area of the XY body, but is also enriched at the unsynapsed axes. Inhibition of pre-rRNA biogenesis abolishes phase separation of DSB repair factors (aka IRIF and the XY body), suggesting that pre-rRNA is one of the major components to maintain the phase separation of DSB repair factors.
Two different methods of fixation were engaged to examine the localization of pre-rRNA in the XY body. With paraformaldehyde, most proteins are preserved in the meiotic spreads, and we could observe protein subunits of pre-rRNP co-localizing with DSB repair factors in the XY body. However, due to protein interference, pre-rRNA hybridization in the XY body is compromised, and its localization at the unsynapsed axes was not very clearly shown under this fixation condition. Thus, we chose methanol-acidic acid to fix the meiotic spreads, which removed proteins and allowed us to clearly observe the string-like pattern of pre-rRNA. For those pre-rRNAs, we only found that 34S and 32S pre-rRNA in the XY body, but not their ancestor 45S pre-rRNA. It is because 45S pre-rRNA is quickly processed once it is transcribed from rDNA loci. It has been estimated that the half-life of 45S pre-rRNA is merely 30 min in cell.50,51,52 Comparing the signal intensity of RNA probes in the XY body, we roughly calculated the rRNA components in the XY body, which are very similar to that in nucleolus. In fact, we observed pre-rRNA infiltration into the XY body from early pachytene to diplotene, suggesting a possible fusion of nucleolus and the XY body. Moreover, like pre-rRNA, other nucleolus components are also detected in the XY body, further indicating the penetration of nucleolus components into the XY body. We failed to find mature rRNA in the XY body, or other translation machinery at DNA lesions, suggesting that these pre-rRNAs may not be involved in protein translation.
We have demonstrated that pre-rRNA is recognized by various repair factors at DNA lesions. Among these repair factors, MDC1 also plays a key role in maintaining phase separation of DSB repair factors in both XY body and IRIF.43,44,53 We found that both the FHA domain and the PST repeats of MDC1 interacted with pre-rRNA. It is possible that the interaction between MDC1 and pre-rRNA is the molecular basis for the phase separation of DSB repair factors. Lacking either of them abolished the phase separation (aka the XY body and IRIF) (Supplementary information, Fig. S17). In MDC1-null cells, γH2AX could not form distinguished foci and γH2AX condensates had a fuzzy boundary (Fig. 6d and Supplementary information, Fig. S9e). Consistently, without the pre-rRNA-associated FHA domain or the PST repeats, MDC1 failed to maintain clear γH2AX foci (Supplementary information, Fig. S16a). Moreover, pre-rRNA transcription suppression by pol I inhibitor also induces similar phenomenon (Fig. 7a and Supplementary information, Fig. S12a). Collectively, these results further indicate that the interaction between MDC1 and pre-rRNA likely provides scaffold for the XY body and IRIF formation, aka the phase separation of DSB repair factors. Consistently, a recent study indicates that the PST repeats of MDC1 are critical to attract HR factors to DSBs, perhaps independent of H2AX.53 In addition to MDC1, other repair factors such as BRCA1 and TOPBP1 also recognize pre-rRNA in the XY body and IRIF. Since these repair factors are downstream of MDC1 during IRIF formation, it is likely that the MDC1/pre-rRNA complex facilitates the recruitment of these repair factors to DNA lesions (Supplementary information, Fig. S17). In BRCA1 and TOPBP1, the BRCT domain recognizes pre-rRNA. It has been shown that the BRCT domain recognizes phospho-Ser motifs.54,55 However, none of the known protein-binding partners is required for the recruitment of BRCA1 and TOPBP1. Interestingly, nucleic acid such as pre-rRNA contains large amount of phosphate moieties. Thus, it is possible that a set of BRCT domain recognizes phosphate groups in pre-rRNA. Further analysis on these BRCT domains may reveal detailed interactions between DSB repair factors and their functional partners in the phase separation.
Pre-rRNAs associate with proteins and other non-coding RNAs to form pre-RNPs.56 Here, we also found these proteins and RNAs exist in the XY body and IRIF, while their functions remain elusive in the XY body and IRIF. It is possible that these proteins and RNAs act as passengers to be loaded at DNA lesions by pre-rRNAs. Alternatively, different proteins and RNAs may have unique functions in the XY body and IRIF. In particular, some protein partners exist in the whole XY body, while others localize on the unsynapsed axes, suggesting that different forms of pre-rRNPs may play different roles in DSB repair. Nevertheless, this study opens a new avenue for future exploring DSB repair.
In addition to phase separation, pre-rRNA may have other profound functions at DSB sites. In particular, pre-rRNA is enriched and colocalizes with RAD51 at the unsynapsed axes. Loss of MDC1 does not completely abolish the association of pre-rRNA onto the X chromosome during meiotic prophase, indicating that in addition to MDC1, other partners of pre-rRNA may mediate this association. Moreover, recent studies have shown that a number of ribosome protein subunits are substrates of PARP1.57,58,59,60,61 Given the role of PARP1 in DDR, it is possible that ADP-ribosylation of these ribosome protein subunits may play key roles in DNA damage repair. We have shown that poly-ADP-ribose serves as DNA damage signals for mediating the recruitment of numerous repair factors to DNA lesions.62,63,64,65,66 And this is the first wave of signals in response to DNA damage.67,68 It is possible that ribosome protein subunits are PARylated and serve as platform for host repair factors at DNA lesions.
Materials and methods
Plasmids
DNA fragment encoding the BRCA1-BRCT domain, TopBP1-BRCT5 domain, MDC1-FHA domain, MDC1 BRCT domain or MDC1-PST repeats was cloned into the pGEX-4T1 vector for recombinant protein purification. DNA fragment encoding full-length, ΔFHA or ΔPST mutant MDC1 was cloned into SFB vector for reconstitution of MDC1 in MDC1-deificent cells.
Animals
All mice experiments were permitted by Westlake University Animal Care and Use Committee and were performed according to the approved protocol. WT mice were obtained from Westlake University Animal Center. H2ax−/−, Mdc−/− and Rnf8−/− mice were gifted by Dr Linyu Lu (Zhejiang University).
Antibodies
The following antibodies were purchased from respective companies: Anti-rRNA (Novus, NB100‐662), anti-RPL7A (ABclonal, A13713), anti-RPL6 (Proteintech, 15387-1-AP), anti-RPS3 (ABclonal, A2533), anti-FBL (CST, 2639S), anti-RPL4 (Proteintech, 11302-1-AP), anti-RPL13 (Proteintech, 11271-1-AP), anti-RPL14 (Proteintech, 14991-1-AP), anti-SCP3 (Santa Cruz, sc-74569; Novus, NB300-232), anti-TopBP1 (Santa Cruz, sc-271043), anti-MDC1 (Abcam, ab11169), anti-53bp1 (Abcam, ab172580), and anti-DNA-RNA Hybrid, clone S9.6 (Millipore, MABE1095).
Surface spread of spermatocytes (paraformaldehyde fixation)
Tunica albuginea was first removed from the tests. The seminiferous tubules were then incubated in buffer I (30 mM Tris-HCl, pH 8.2, 50 mM sucrose, 17 mM sodium citrate, 5 mM EDTA, 0.5 mM dithiothreitol and 0.1 mM phenymethylsulfonyl fluoride) for 15 min at room temperature. Spermatocytes were released to the buffer I by forceps and same volume buffer II (100 mM sucrose) was added. This mixture was then screened by a 70 μm cell strainer. Finally, the screened cells were spread onto buffer III (1% PFA, 0.15% Triton X-100, pH 9.2) pre-soaked glass slides. These slides were kept in humidified chambers and air dried overnight.
Surface spread of spermatocytes (methanol/acetic acid fixation)
The seminiferous tubules were incubated in KCL solution (75 mM) for 1 h at room temperature. Then the spermatocytes were released by forceps and screened by a 70 μm cell strainer. Subsequently, 8 mL mixture was added with 1 mL freshly prepared Methanol/Acetic Acid fixation fixative (Methanol:Acetic Acid = 3:1) and kept at room temperature for 10 min. Spermatocytes were spinned down and resuspended with fixative and incubated for 20 min at room temperature. After incubation, the cells were spinned down and resuspended with moderate volume of fixative. Finally, the cells were dropped onto glass slides and air dried.
RNA fluorescence in situ hybridization (RNA FISH) and immunofluorescence (IF) staining for meiotic spreads
Meiotic spreads were first incubated with wash buffer A (SMF-WA1-60, Bioresearch technologies, USA) for 5 min at room temperature. Slides were then incubated with probes diluted by hybridization buffer (SMF-HB1-10, Bioresearch technologies, USA) overnight at 37 °C. After probes incubation, the slides were washed by wash buffer A for 10–30 min at room temperature. All the probes are listed in Supplementary Table S1. To detect pre-rRNA, we mainly used probe 4 and 6 for in situ hybridization.
For IF staining, slides were incubated with primary antibodies diluted with 1% BSA for 2–3 h at room temperature or overnight at 4 °C. PBS was subsequently used to wash the slides three times. The slides were then incubated with corresponding secondary antibodies for 2 h at room temperature and washed three times with PBS. Hoechst staining (10 min at room temperature) was done after RNA FISH or IF staining. Of note, the IF staining was performed after RNA FISH staining in detection of both RNAs and proteins.
Harvesting spermatogonia
In the testis of 3-week-old mice, most spermatogonia are in pachytene stage. Tunica albuginea was first removed from the tests of 3-week-old mice. Spermatocytes were released to DMEM medium by forceps. This mixture was then screened by a 70 μm cell strainer. The cells were then cultured in DMEM medium supplemented with 10% FBS and 1% penicillin and streptomycin at 37 °C with 5% CO2. One hour later, most cells in the supernatant were germ cells and were collected for RNA-ChIP assay.
RNA-ChIP assay
Germ cells were fixed in 1% formaldehyde for 10 min at room temperature, and 0.125 M glycine was used to quench cross-linking with for 5 min. Cells were washed twice with cold PBS and lysed with RIP buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.5, 0.5% NP-40, 1 mM DTT, 100 U/mL RNase inhibitor, protease inhibitor cocktail). The cells are homogenized by sonication to an average fragment size of 1000 nucleotides and were centrifuged at 15,000 rpm for 30 min at 4 °C to remove the insoluble materials. Next, lysis was pre-cleared with protein A/G beads in cold room and immunoprecipitated with primary antibody following the ChIP protocol. The beads were washed, and crosslinks were reversed following the ChIP protocol. Then, RNA was precipitated with phenol-chloroform and treated extensively with DNase I. After ethanol-precipitated, the RNA was resuspended with nuclease free water. RNA-seq was performed by LC-Bio Technology CO. Ltd., Hangzhou, China, with GEO number GSE184711.
RNA and GST-protein pull-down assay
Purified N-terminal GST-tagged proteins were incubated with total RNA and Glutathione SepharoseTM 4B (17-0756-05, GE Healthcare) at 4 °C for 2 h. The reaction mixture was washed by NETN100 three times at 4 °C, and the supernatant was discarded. PK buffer was then added to the sediment, which was mixed and incubated for 10 min at 37 °C. Same volume of Urea/PK buffer was subsequently added to the mixture and incubated for another 10 min at 37 °C. After incubation, the mixture was centrifuged at 4 °C and the supernatant was discarded. The prepared RNA precipitator (1 mL ethanol, 40 μL 3 M sodium acetate and 1 μL glycogen) was used to precipitate RNA at –80 °C overnight. Finally, the protein-binding RNA was purified by centrifugation (13,000 rpm 10 min at 4 °C). RT-qPCR was performed to detect the purified RNA. The primers are listed in Supplementary Table S1.
Hybridization of recombinant proteins to meiotic spreads
N-terminal GST-tagged recombinant proteins were diluted in PBS and were incubated with meiotic spreads for 1 h at 37 °C. These slides were then washed by PBS three times. IF staining was performed subsequently. For removing RNA, meiotic spreads were treated with 1 μg/mL RNase A for 30 min at 37 °C. Excessive pre-rRNA extracted from mouse embryo fibroblasts (MEFs) was added to the recombinant proteins solutions for pre-rRNA blocking before hybridization.
Polysome profiling
Cells were first treated with vehicle (DMSO) or BMH-21(1 μM). Before collection, cells were treated with 100 μg/mL cycloheximide (CHX) for 10 min. Cells were washed three times with PBS supplemented with 100 μg/mL CHX, then centrifuged at 1000 rpm for 3 min at 4 °C. Subsequently, 500 μL lysis buffer (25 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 150 mM NaCl, 1 mM DTT, 1% Triton X-100, 0.5% Na-Deoxycholate, 8% Glycerol, 100 μg/mL CHX, 100 U/mL SUPERase inhibitor) was used to resuspend the cells. After centrifugation at 12,500 rpm for 10 min, the precipitate containing debris and nuclei was discarded. Total RNA was evaluated by A260 and 7.5 OD of lysates were loaded on 10%–50% sucrose gradients. The sucrose gradients were centrifuged on a SW41Ti rotor (Beckman Coulter) for 4 h at 40,000 rpm at 4 °C. The fractioned samples were measured by Microplate reader (Varioskan LUX).
Protein synthesis measurement
The protein synthesis of vehicle (DMSO)- or BMH-21-treated cells was measured by the incorporation of puromycin into peptide chains. In brief, cells were first treated with vehicle (DMSO) or BMH-21 (1 μM) and subsequently incorporated with puromycin for 10 min. Then the cells were collected for western blotting assay. Anti-puromycin antibody (clone 12D10, Merk, MABE343) was applied to quantitate overall neosynthesized protein.
BMH-21 treatment in mice
Ten 3-week-old male mice were separated to two groups, the vehicle (DMSO) or BMH-21 (40 mg/kg) treated group. DMSO or BMH-21 were injected every 3 days. Mice were sacrificed after 3 weeks’ treatment. The testes were removed from the mice and were used for the following analysis.
ChIP in I-SceI-induced DSB system
An I-SceI site was knockedin on the X chromosome in HCT116 cells. HA-I-SceI-GR pCW tet-on plasmid was transfected into the cells. Doxycycline was applied to induce I-SceI expression and acetonide (TA) was applied to translocate I-SceI to the nucleus. Finally, I-SceI cut the I-SceI site and caused DSB. ChIP-qPCR was performed to determine the occupation of rRNA, FBL, γH2AX, MDC1, BRCA1 or TopBP1 in this specific DSB site. Primers are listed in Supplementary Table S1.
GFP-reporter assay to detect the efficiency of homologous recombination repair
DR-GFP reporter is designed to measure homology-directed repair (HDR), where a gene fragment (iGFP) serves as a template for HDR of an I-SceI-induced DSB in an upstream SceGFP cassette. HA-I-SceI-GR pCW tet-on plasmid was transfected to the cells. Doxycycline was used to induce I-SceI expression and acetonide (TA) was applied to translocate I-SceI to the nucleus. Finally, I-SceI cut the I-SceI site and caused DSB. Once this DSB was repaired, the GFP expressed. Flow cytometry was performed to examine the efficiency of homologous recombination repair.
Detection of rRNA and pre-rRNA-associated proteins in IRIF
Cells were plated on glass coverslips and treated with IR. After washing with PBS, the cells were treated with HEB buffer for 30 min and then with sucrose solution (100 mM) for 30 min. The cells were fixed with 2% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS for 30 min at room temperature. Stellaris RNA FISH and immunofluorescence were performed according to the manufacturer’s protocol. Pre-rRNA probes are listed in Supplementary Table S1.
Biostatistical visualization
Analysis of pre-rRNA and SCP3 on X and Y chromosomes: the 100 foci showing highest signal strength were extracted together with the neighbor data (±1 μm) from the immunofluorence photograph. The normalized signal strength was visualized using ComplexHeatmap.
γH2AX foci analysis: the immunofluorescence photographs were processed using python-based artificial detection algorithm to fetch loci with highest response signals (correlated with the absolute value on the green channel of the photograph, while the value ranged from 0 to 255). This algorithm was optimized with a radius threshold of 0.5 μm so that the selected loci would not affect each other. After extracting the co-ordinate of the top 20 loci of each photograph with their absolute value on the green channel, we used an R script to determine which axis (horizontal or vertical) would be selected for each locus, based on the comparison of standard deviation with the loci’s neighborhood within a radius of 0.5 μm. The absolute values extracted from the green channel were divided by 255 to get a normalized matrix and then outputted as a heatmap using ComplexHeatmap.
In vitro phase separation and RNA buffering assay
For the in vitro LLPS experiments, purified FHA or PST protein was mixed with the phase separation buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.4 and 10% PEG 8000 (NEB)) and was directly pipetted onto a coverslip under the microscope. During the LLPS experiments, every dynamic process was observed by a Nikon microscope with 40× differential interference contrast (DIC).
For the RNA buffering assay, the proteins were incubated with pre-RNAs in the above phase separation buffer. Finally, 10 μL of each sample was pipetted onto a coverslip and imaged. The pre-RNAs from 293T cells were separated by agarose electrophoresis and collected by Spin Column RNA Cleanup & Concentration Kit (Sangon Biotech).
Statistical analysis
Data were analyzed with GraphPad Prism 7.0 software. The data presented were means ± SD. Statistical significance between two groups was subjected to Student’s two-tailed t-test. P < 0.05 was considered statistically significant.
References
Harper, J. W. & Elledge, S. J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Lukas, J., Lukas, C. & Bartek, J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169 (2011).
Yuan, J., Adamski, R. & Chen, J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 584, 3717–3724 (2010).
Paull, T. T. et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 (2000).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675–679 (2003).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Celeste, A. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003).
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
Neale, M. J., Ramachandran, M., Trelles-Sticken, E., Scherthan, H. & Goldman, A. S. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell 9, 835–846 (2002).
Romanienko, P. J. & Camerini-Otero, R. D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000).
Keeney, S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53 (2001).
Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Lu, L. Y. & Yu, X. Double-strand break repair on sex chromosomes: challenges during male meiotic prophase. Cell Cycle 14, 516–525 (2015).
Lu, L. Y., **ong, Y., Kuang, H., Korakavi, G. & Yu, X. Regulation of the DNA damage response on male meiotic sex chromosomes. Nat. Commun. 4, 2105 (2013).
Haaf, T., Golub, E. I., Reddy, G., Radding, C. M. & Ward, D. C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA. 92, 2298–2302 (1995).
Carofiglio, F. et al. SPO11-independent DNA repair foci and their role in meiotic silencing. PLoS Genet. 9, e1003538 (2013).
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M. & Lamond, A. I. The nucleolus under stress. Mol. Cell 40, 216–227 (2010).
Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021).
Wei, W. et al. A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112 (2012).
Yamanaka, S. & Siomi, H. diRNA-Ago2-RAD51 complexes at double-strand break sites. Cell Res. 24, 511–512 (2014).
Shadrina, O. et al. Analysis of RNA binding properties of human Ku protein reveals its interactions with 7SK snRNA and protein components of 7SK snRNP complex. Biochimie 171-172, 110–123 (2020).
Lee, S. et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016).
Munschauer, M. et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 561, 132–136 (2018).
Michelini, F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017).
D’Alessandro, G. et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 9, 5376 (2018).
Hu, Y. et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 32, 2250–2262 (2018).
Cowell, I. G. et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111, 22–36 (2002).
Sanz, L. A. & Chedin, F. High-resolution, strand-specific R-loop map** via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734–1755 (2019).
Mullineux, S. T. & Lafontaine, D. L. Map** the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 94, 1521–1532 (2012).
Karijolich, J., Kantartzis, A. & Yu, Y. T. RNA modifications: a mechanism that modulates gene expression. Methods Mol. Biol. 629, 1–19 (2010).
Zhang, J. et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592 (2007).
Kressler, D. et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 338, 666–671 (2012).
Peculis, B. A. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA 7, 207–219 (2001).
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005).
Lou, Z. et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 (2006).
Ichijima, Y. et al. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 25, 959–971 (2011).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Lu, L. Y. et al. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 18, 371–384 (2010).
Yang, G. et al. Poly(ADP-ribosyl)ation mediates early phase histone eviction at DNA lesions. Nucleic Acids Res. 48, 3001–3013 (2020).
Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021).
Popov, A. et al. Duration of the first steps of the human rRNA processing. Nucleus 4, 134–141 (2013).
Hall, T. J. & Cummings, M. R. In vitro synthesis and processing of ribosomal RNA in the housefly ovary. Dev. Biol. 46, 233–242 (1975).
Eliceiri, G. L. Short-lived, small RNAs in the cytoplasm of HeLa cells. Cell 3, 11–14 (1974).
Salguero, I. et al. MDC1 PST-repeat region promotes histone H2AX-independent chromatin association and DNA damage tolerance. Nat. Commun. 10, 5191 (2019).
Yu, X., Chini, C. C., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Botuyan, M. V. et al. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure 12, 1137–1146 (2004).
Perez-Fernandez, J., Roman, A., De Las Rivas, J., Bustelo, X. R. & Dosil, M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 27, 5414–5429 (2007).
Challa, S. et al. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell 184, 4531–4546, e4526 (2021).
Kim, D. S. et al. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell 75, 1270–1285.e1214 (2019).
Boamah, E. K., Kotova, E., Garabedian, M., Jarnik, M. & Tulin, A. V. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 8, e1002442 (2012).
Zhen, Y., Zhang, Y. & Yu, Y. A cell-line-specific atlas of PARP-mediated protein Asp/Glu-ADP-ribosylation in breast cancer. Cell Rep. 21, 2326–2337 (2017).
Leitinger, N. & Wesierska-Gadek, J. ADP-ribosylation of nucleolar proteins in HeLa tumor cells. J. Cell Biochem. 52, 153–158 (1993).
Hou, W. H., Chen, S. H. & Yu, X. Poly-ADP ribosylation in DNA damage response and cancer therapy. Mutat. Res. Rev. Mutat. Res. 780, 82–91 (2019).
Li, M., Lu, L. Y., Yang, C. Y., Wang, S. & Yu, X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 27, 1752–1768 (2013).
Li, M. & Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013).
Zhang, F., Chen, Y., Li, M. & Yu, X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. USA 111, 7278–7283 (2014).
Zhang, F., Shi, J., Chen, S. H., Bian, C. & Yu, X. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 43, 10782–10794 (2015).
Haince, J. F. et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008).
Liu, C., Vyas, A., Kassab, M. A., Singh, A. K. & Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 (2017).
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (32090034 and 81874160), Westlake University Education Foundation and Westlake Laboratory of Life Sciences and Biomedicine. We thank Dr Linyu Lu (Zhejiang University) for sharing H2ax−/−, Mdc1−/− and Rnf8−/− mice. We also thank Dr Peiguo Yang for phase separation analyses and Guicun Fang for the image analysis. We thank Dr Junyi Lin (School of Basic Medical Sciences, Fudan University) for the help of data analysis. We appreciate the supports of Microscope Core Facility and Animal Center of Westlake University.
Author information
Authors and Affiliations
Contributions
X.Y. conceived and designed the project. X.G., D.X., D.W., X.W., L.C., Y.W., K.M. and P.L. performed the experiments. X.Y. wrote the manuscript. X.G. organized figures and edited the manuscript. Q. L. analyzed the RNA-seq data and some of the IF images. All authors read and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Gai, X., **n, D., Wu, D. et al. Pre-ribosomal RNA reorganizes DNA damage repair factors in nucleus during meiotic prophase and DNA damage response. Cell Res 32, 254–268 (2022). https://doi.org/10.1038/s41422-021-00597-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-021-00597-4
- Springer Nature Singapore Pte Ltd.
This article is cited by
-
A graph neural network-based interpretable framework reveals a novel DNA fragility–associated chromatin structural unit
Genome Biology (2023)
-
RPLP2 activates TLR4 in an autocrine manner and promotes HIF-1α-induced metabolic reprogramming in hepatocellular carcinoma
Cell Death Discovery (2023)
-
Liquid-liquid phase separation in DNA double-strand breaks repair
Cell Death & Disease (2023)
-
Comparative transcriptome analysis to unveil genes affecting the host cuticle destruction in Metarhizium rileyi
Current Genetics (2023)