Abstract
Pyroptosis is a form of cell death triggered by the innate immune system that has been implicated in the pathogenesis of sepsis and acute lung injury. At the cellular level, pyroptosis is characterized by cell swelling, membrane rupture, and release of inflammatory cytokines, such as IL-1β. However, the role of endogenous lipids in pyroptosis remains underappreciated. We discovered that 4-hydroxynonenal (HNE), a major endogenous product of lipid peroxidation, inhibited pyroptosis and inflammasome activation. HNE at physiological concentrations (3 µM) blocked nigericin and ATP-induced cell death, as well as secretion of IL-1β, by mouse primary macrophages and human peripheral blood mononuclear cells. Treatment with HNE, or an increase of endogenous HNE by inhibiting glutathione peroxidase 4, reduced inflammasome activation in mouse models of acute lung injury and sepsis. Mechanistically, HNE inhibited the NLRP3 inflammasome activation independently of Nrf2 and NF-κB signaling, and had no effect on the NLRC4 or AIM2 inflammasome. Furthermore, HNE directly bound to NLRP3 and inhibited its interaction with NEK7. Our findings identify HNE as a novel, endogenous inhibitor of the NLRP3 inflammasome.
Similar content being viewed by others
Introduction
Pyroptosis is a lytic form of programmed cell death, characterized by cytoplasmic swelling, pore formation in the cell membrane, and release of pro-inflammatory cytokines [1]. Pyroptosis is initiated in response to pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) via pattern recognition receptors (PRRs) [2]. Based on their location, PRRs are divided into membrane-bound PRRs and cytoplasmic PRRs. Toll-like receptors (TLRs) are transmembrane proteins that play important roles in the innate immune response [3]. For example, activation of the TLR4 receptor by gram-negative bacteria endotoxins, such as lipopolysaccharide (LPS), stimulates multiple signaling pathways in macrophages, including NF-κB, and the subsequent production of pro-inflammatory cytokines [3]. Unlike TLRs, the nucleotide-binding oligomerization domain-like receptors (NOD-like receptors, NLRs) recognize endogenous danger or stress responses, and form multiprotein complexes called inflammasomes [4,5,6]. NLR Family Pyrin Domain Containing 3 (NLRP3), NLR family CARD domain-containing protein 4 (NLRC4), NLR Family Pyrin Domain Containing 1 (NLRP1) and Absent In Melanoma 2 (AIM2) are the best characterized inflammasomes, and have been implicated in the pathogenesis of sepsis, atherosclerosis, and acute lung injury [4,5,6,7]. Stimulation of inflammasomes involves two signals: 1) transcriptional and posttranslational priming of inflammasome components, for example by LPS; and 2) activation of inflammasome assembly by a cell danger signal, such as K+ influx, extracellular ATP for NLRP3, or double-stranded DNA for AIM2. The formation of inflammasomes triggers the activation of caspase-1 and subsequent processing of interleukin-1β (IL-1β) and interleukin-18 (IL-18) into their mature forms [8, 9]. Gasdermin-D (GSDMD) was discovered as a pore-forming protein and the key effector downstream of caspase-1 activation. Active caspases cleave GSDMD to generate an N-terminal cleavage product (GSDMD-NT) that forms transmembrane pores to enable IL-1β release and to drive pyroptosis [10,11,12]. The importance of IL-1β as a disease mediator was confirmed by the CANTOS trial in which an IL-1β neutralizing antibody led to a lower rate of recurrent cardiovascular events in patients with previous myocardial infarction [13].
Recent data suggest that endogenous lipids or their oxidation products can activate or inhibit the assembly of inflammasomes [5, 14]. Among reactive aldehydes derived from lipid peroxidation, 4-hydroxynonenal (HNE) is the most abundant end-product. The concentration of HNE in human serum is 0.05–0.15 μM under physiological conditions [15]. However, HNE levels may reach 3–6 μM in tissues under oxidative stress [16, 17]. Because of its high solubility in aqueous fluids, the reactive HNE formed in membranes can diffuse into the cytoplasm. HNE is detoxified by conjugation to glutathione by glutathione S-transferase [18, 19]. However, some HNE molecules escape this mechanism and react with the side chains of cysteine, histidine and lysine residues in proteins [20,21,22]. HNE thus has emerged as an important second messenger signaling molecule [18, 19]. For example, low concentrations of HNE produce beneficial effects, including the stimulation of endogenous antioxidant defense mechanisms and the inhibition of inflammation [19, 23,24,25,26]. Currently, two mechanisms are proposed for HNE-mediated regulation of inflammation. (1) HNE facilitates antioxidant expression by activating Nrf2 signaling, via disrupting Keap1 − Nrf2 association and preventing Nrf 2 degradation [24, 25, 27]. Nrf2 stimulates antioxidant expression and increases the resistance to cytotoxic reactive oxygen species (ROS), thereby blocking multiple inflammatory pathways. (2) HNE blocks NF-κB activation by inhibiting IκB kinase (IKK) activity, likely by covalently modifying cysteine residue(s) of IKK [26, 28]. In this study we explored a novel mechanism by which the lipid peroxidation product HNE inhibits the NLRP3 inflammasome by directly disrupting the binding of NEK7 to NLRP3, thereby preventing activation of caspase-1. We demonstrate the importance of this pathway in decreasing inflammatory cytokine release and macrophage pyroptosis in vitro and in vivo.
Results
HNE inhibits pyroptotic cell death in human and mouse macrophages
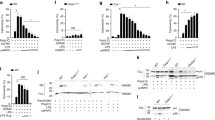
Nigericin is a K+ ionophore that activates the NLRP3 inflammasome [29] and stimulates pyroptotic cell death. To study the role of HNE in pyroptosis, we used LPS co-incubated with HNE, followed by nigericin, in differentiated human THP-1 macrophages and mouse bone-marrow-derived macrophages (BMDMs). Cell death was confirmed by morphological changes, lactate dehydrogenase (LDH) release, and real-time nucleic acid staining (SYTOXTM). HNE treatment alone (3 µM) had no effect on cell death, but HNE significantly decreased the magnitude of LPS-nigericin stimulated cell death as indicated by membrane blebbing, LDH release, and nucleic acid staining in mouse and human macrophages (Fig. 1A–F). These results show that HNE protects macrophages from LPS/nigericin-mediated pyroptosis.
A–C THP-1 differentiated macrophages were stimulated with LPS (100 ng/mL) and co-incubated with HNE (3 μM) or vehicle (ethanol) for 3 h followed by 2 h of 6 μM nigericin (Nig) treatment. A Cell morphology, scale bar = 10 µm. B LDH cytotoxicity. C Cell death by SYTOXTM green. D–F Bone-marrow-derived macrophages (BMDMs) were stimulated with LPS (100 ng/mL) and co-incubated with HNE (3 μM) or vehicle (ethanol) for 3 h followed by 1 h of 2 μM nigericin (Nig) treatment. D Cell morphology, scale bar = 10 µm. E LDH cytotoxicity. F Cell death by SYTOXTM green. Statistics in B and E were performed using a 2-way ANOVA and Bonferroni’s post hoc test. ****P < 0.001 between LPS + Nig+ethanol and LPS + Nig+HNE groups. (N = 4 experiments). Bars represent mean ± SD.
HNE inhibits pyroptosis independent of Nrf2 signaling
Nrf2, an antioxidant transcription factor, has been proposed to regulate NLRP3 inflammasome activation [30,31,32]. Nrf2 function is inhibited by Keap1, which binds Nrf2, facilitates its degradation, and prevents its nuclear translocation [33]. To study the potential role of Nrf2 in HNE inhibition of pyroptosis, we first showed that HNE (3 µM) induced Nrf2 activation in macrophages as indicated by its nuclear translocation (Fig. 2A, B), wherase LPS did not. We next studied the effect of HNE on Nrf2 regulated genes, such as glutamate-cysteine ligase catalytic subunit (GCLC) and ferroportin-1 (Fig. 2C, D) [34]. LPS treatment alone had no significant effect on any of these measurements (Fig. 2C, D). Simultaneous treatment with LPS and HNE significantly increased Nrf2 activation, compared to LPS alone (Fig. 2A–D). To determine if Nrf2 activation was required for HNE inhibition of pyroptosis, we blocked Nrf2 signaling using the specific inhibitor, ML385 [35] (Fig. S1A, B). We found that 3 µM HNE still prevented cell death in the presence of ML385 (Fig. 2E, F), suggesting that the protective effect of HNE against pyroptosis was independent of the Nrf2 pathway.
A, B THP-1 macrophages were stimulated with or without LPS (100 ng/mL) and co-incubated with ethanol or 3 µM HNE for 3 h. A Nrf2 translocation was determined by immunofluorescence (scale bar: 10 µm), DAPI (blue), and Nrf2 (Green). B Nuclear to cytosolic Nrf2 intensity ratio was quantified by Image J. C, D Peritoneal macrophages were stimulated with or without LPS (100 ng/mL) and co-incubated with ethanol or 3 µM HNE for 3 h. Gene expression was analyzed by real-time PCR. C GCLC mRNA expression and (D) Ferroportin-1 mRNA expression were measured after normalizing to β-actin expression. E, F THP-1 macrophages were treated with LPS with or without 3 µM HNE or 2 µM ML385 for 3 h followed by 6 µM nigericin (Nig) to induce inflammasome activation. E Cell morphology, scale bar = 5 µm. F Cell death by SYTOXTM green. Statistics in B, C, and D were performed using a 2-way ANOVA and Bonferroni’s post hoc test. **P < 0.01, ****P < 0.001 between HNE and ethanol groups after LPS treatment. Bars represent mean ± SD.
HNE inhibits inflammasome activation independently of NF-κB signaling
Activation of the inflammasome involves first the NF-κB dependent stimulation of NLRP3 expression, and then NLRP3 oligomerization promoted by a cell danger signal, such as K+ efflux or ATP [36]. We first focused on the role of HNE in NF-κB signaling by assessing p65 phosphorylation, p65 nuclear translocation, and IκB-α degradation, as well as the expression of TNF-α and NLRP3 in macrophages treated with LPS. In response to LPS (100 ng/mL), there was a 3-fold increase in nuclear translocation of p65 (Fig. 3A, B) and a 10-fold increase in phosphorylation of p65 (Fig. 3C). TNF-α and NLRP3 expression were induced after LPS treatment through the NF-κB pathway, but these measurements were not significantly affected by 3 µM HNE (Fig.3A–C, Fig. S2A, B). Similarly, there was no significant effect of HNE (0.3–3 µM) on NLRP3, pro-IL-1β, and IκB-α protein expression (Fig. 3D–F, and Fig. S2D). Interestingly, HNE significantly reduced pro-IL-1β gene expression (Fig. S2C), suggesting that longer treatment of HNE may affect pro-IL-1β protein expression. Inflammasome activation induced by nigericin triggers IL-1β cleavage in LPS-primed macrophages. To minimize the effects of HNE on NF-κB-dependent transcription, macrophages were treated with LPS for 3 h before exposure to HNE (Fig. S2E) and then stimulated with nigericin (Fig. 3G). We found HNE still significantly prevented IL-1β maturation (Fig. 3H). Furthermore, we performed the experiments where BMDMs were pre-treated with HNE for 30 min, and then stimulated with nigericin after 10 min LPS priming. NLRP3 activation was confirmed by the cleavage of caspase-1 and GSDMD. We found that rapid NLRP3 priming followed by nigericin treatment induced caspase-1 and GSDMD cleavage, and HNE pre-treatment inhibited these effects (Fig. S3). Thus, the inhibitory effect of HNE treatment on inflammasome activation is independent of the NF-κB pathway.
A, B THP-1 macrophages were stimulated with or without LPS (100 ng/mL) and co-incubated with ethanol or 3 µM HNE for 1 h. A NF-κB p65 translocation was measured by immunofluorescence (scale bar: 5 µm), DAPI (blue), and p65 (Green). B Quantification using Image J. N = 3 experiments. C–E Peritoneal macrophages were stimulated with or without LPS (100 ng/mL) and co-incubated with ethanol or HNE (0.3-3 µM) for 3 h. Protein expression was analyzed by western blot. C Phosphorylation of p65, D NLRP3, E Pro-IL-1β. Bars represent mean ± SEM. N = 3 experiments. F Peritoneal macrophages were stimulated with LPS (100 ng/mL) and co-incubated with ethanol or HNE (3 µM) for 1or 3 h. Cell lysates were analyzed by western blot. F NLRP3, IκB-α, and β-actin western blots are representative of three independent experiments. G Schematic of experimental design for data in Fig. 3H. H BMDMs were stimulated with LPS (100 ng/mL) for 3 h followed by 6 µM nigericin (Nig) for 30 min. 3 µM HNE or ethanol was added 30 min before nigericin. Western blots (NLRP3, p65 phosphorylation, β-actin, pro-IL-1β, and cleaved IL-1β) are representative of three independent experiments. Statistics in B were performed using a 2-way ANOVA and Bonferroni’s post hoc test. Statistics in C–E were performed using a one-way ANOVA and Bonferroni’s post hoc test. N = 3 experiments. A p-value less than 0.05 is statistically significant among treatment groups. Bars represent mean ± SD.
HNE inhibits NLRP3 inflammasome activation, but has no effect on AIM2 or NLRC4
We next studied the effect of HNE on inflammasome assembly and activation. It has been suggested that NLRP3 and apoptosis-associated speck-like (ASC) proteins are assembled by acetylated α-tubulin-mediated transport to the microtubule organizing center (MTOC) [37]. Upon inflammasome activation by nigericin stimulated potassium efflux, we found increased α-tubulin acetylation in LPS primed peritoneal macrophages (Fig. 4A), BMDMs (Fig. S4A), and THP1 macrophages (Fig. S4B). However, HNE had no effect on acetylation of α-tubulin in these cells (Fig. 4A and Fig. S4A, B). These data suggest that acetylated α-tubulin was not involved in HNE inhibition of pyroptosis.
A–F HNE (3 µM) or ethanol was added 30 min before nigericin, ATP, or poly(dA:dT) treatment. A Peritoneal macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by 2 µM nigericin (Nig). Acetyl-α-Tubulin (Lys40) western blots are representative of three independent experiments. B Peritoneal macrophages. C Human PBMC were stimulated with LPS (100 ng/mL) for 3 h followed by 2 µM nigericin (Nig) or ATP (2 mM) stimulation for 1 h. IL-1β in the medium was measured by ELISA. Bars represent mean ± SD. D THP-1 macrophages that overexpressed ASC-GFP were stimulated with LPS (100 ng/mL) followed by 6 µM nigericin (Nig) for 2 h. ASC speck formation (arrows) was measured by confocal microscopy. (scale bar: 10 µm). Quantification results were shown at lower magnification images in Fig. S6. E THP-1 macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by 6 µM nigericin (Nig) for 2 h. ASC oligomerization western blots are representative of three independent experiments. F Peritoneal macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by 2 µM nigericin (Nig) or ATP (2 mM) stimulation for 15 min. Western blots are representative of three independent experiments. G Peritoneal macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by 2 µg/mL Poly(dA:dT) for 6 h. Ethanol or 3 µM HNE was co-incubated with cells 30 min before Poly(dA:dT). Western blots are representative of three independent experiments. H THP-1 macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by 2 µg/mL Poly(dA:dT) for 6 h. IL-1β in the medium was measured by ELISA. Statistics in B, C, and H were performed using a 2-way ANOVA and Bonferroni’s post hoc test. ****P < 0.001 between control and treatment groups. Bars represent mean ± SD.
To analyze the effects of HNE more broadly, we used ATP as a second signal to induce inflammasome activation (Fig. 4B, C). We also studied both peripheral blood mononuclear cells (PBMC) from healthy human donors, and peritoneal macrophages from mice. We confirmed that HNE inhibited IL-1β release (Fig. 4B, C). Next, the effect of HNE on a non-potassium efflux dependent NLRP3 stimulus was tested. THP-1 macrophages were stimulated with LPS for 3 h followed by R837 (Imiquimod) for 1 h. We found that HNE reduced IL-1β secretion (Fig. S5). These data indicated that HNE also inhibits non-potassium efflux-mediated NLRP3 activation.
Upon NLRP3 inflammasome activation, the adaptor protein ASC is recruited by NLRP3 and forms large multimeric complexes, termed ASC specks [38]. To determine the effect of HNE on ASC speck formation, we overexpressed an ASC-GFP fusion protein in THP-1-differentiated macrophages and stimulated them with LPS followed by nigericin. HNE treatment inhibited ASC speck formation, as indicated by reduced ASC speck immunofluorescence (Fig. 4D, and Fig. S6A, B). Similarly, HNE reduced large multimeric ASC complexes, as detected by western blot (Fig. 4E).
Inflammasome activation leads to the cleavage of pro-caspase-1 to generate active caspase-1 that cleaves gasdermin-D (GSDMD) to form membrane pores, which enables the release of cytokines, such as IL-1β. Therefore, we measured the effect of HNE on these parameters of inflammasome activation (Fig. 4F). LPS primed peritoneal macrophages were treated with HNE followed by nigericin or ATP. The inhibitory effect of HNE on NLRP3 activation was confirmed by western blot as shown by decreased cleavage of caspase-1, GSDMD, and IL-1β to their mature p20, p30, and p17 forms, respectively (Fig. 4F).
NLRP3, AIM2, and NLRC4 inflammasomes respond to different ligands or activators, but all engage with the adaptor protein ASC and activate protein caspase-1 to cleave pro-IL-1β. To study the specificity of HNE for the NLRP3 inflammasome, we tested the effect of HNE on the AIM2 and NLRC4 inflammasome. AIM2 is activated by cytosolic double-stranded DNA (dsDNA). NLRC4 is activated by cytosolic flagellin. To determine the effect of HNE on these two inflammasomes, macrophages were treated with LPS for 3 h, then HNE for 30 min, then transfection with poly(dA:dT) or flagellin for 6 h and 3 h, respectively. In contrast to the NLRP3 inflammasome, HNE did not reduce cleavage of caspase-1 or IL-1β release after AIM2 inflammasome activation (Fig. 4G, H). Similarly, HNE did not reduce IL-1β release after NLRC4 inflammasome activation (Fig. S7). Together, these data show that HNE inhibits NLRP3 assembly and activation in mouse and human cells, but has no effect on AIM2 and NLRC4.
HNE reduces non-canonical inflammasome-mediated IL-1β release, but has no effect on GSDMD-mediated cell death
Non-canonical inflammasome activation is dependent on caspase-11 (in mice) or caspase-4 (in humans) which directly bind with intracellular LPS [39, 40]. Recent data suggest that the non-canonical inflammasome pathways play critical roles in acute lung injury and sepsis by sensing cytosolic LPS [Human peripheral blood mononuclear cells (PBMC) isolation PBMC isolation was performed as previously described [85]. Whole blood (20 mL) was layered upon a 15 mL Ficoll-Paque cushion (GE Healthcare, 17-1440-02) in 50 mL conical tubes, and centrifuged for 40 min at 400 x g at room temperature with the brake off. The layer of mononuclear cells at the plasma-density gradient medium interface was transferred to a new 50 mL conical tube, and 1x PBS was added to 45 ml total volume. Cells were centrifuged for 10 min at 2000 x g at room temperature and the remaining pellet of mononuclear cells was resuspended in complete RPMI medium at 1.6 × 105 cells/well in 12-well plates for future experiments. Human THP-1 monocytes were differentiated into macrophages by 24 h incubation with 100 nM PMA (Sigma-Aldrich) in complete RPMI medium at 1.6 × 105 cells/well in 12-well plates. Cells were washed twice with 1x PBS and incubated with complete RPMI medium without PMA for 24 h before experiment. A lentivirus expressing the ASC-GFP fusion protein was prepared by transfection of pLEX-MCS-ASC-GFP (Addgene 73957), psPAX2, and p MD.2 G using Fugene 6 (Promega, PAE2693) in HEK293T cells. Culture medium was replaced after 24 h with fresh medium containing 5% FBS After a further 48 h virus containing medium was collected and stored in −80 °C. THP-1 monocytes at 1.6 × 105 cells/well on 12-well plates were infected with 400 uL virus with polybrene (4 µg/ml) and centrifuged at 2500 rpm at 20 oC for 90 min. Fresh complete RPMI medium with 100 nM PMA was added and followed by 24 h incubation. Cells were washed twice with 1x PBS and incubated with complete RPMI medium without PMA for 24 h before experiment. FLAG-GSDMD-NT (80951) and FLAG-GSDMD-NT-C192A (133891) were obtained from Addgene. HEK293T cells were cultured in complete DMEM medium containing 10% FBS and 1% Penicillin/Streptomycin. Transient transfection of HEK293T cells was performed using Fugene6 (Promega) according to the manufacturer’s instructions. HEK293A cells stably expressing mouse NLRP3 (Addgene 75127), and ASC-GFP (Addgene 73957) were established under G418 (1 mg/ml) and puromycin (1 µg/ml) selection following transfection with Fugene6. NEK7 (Addgene 75142) was expressed in the stable cell line by transient transfection using Fugene6 for 48 h. Cell extracts were prepared after LPS/Nigericin treatment with or without HNE for 30 min. Macrophage cultures were rinsed twice with serum and antibiotic-free medium (RPMI 1640 for peritoneal macrophages, THP-1 macrophages, and PBMCs or DMEM medium for BMDMs). Cells were exposed to LPS (100 ng/mL) for 3 h with ethanol or HNE (0.3–3 µM, Millipore Sigma, 393204) in appropriate growth media. For time-course studies, individual wells were exposed to LPS (100 ng/mL) for different times, but to minimize manipulation of each plate, the total incubation time was 3 h for all treatments. To activate the NLRP3 inflammasome, cells were primed for 3 h with LPS (100 ng/mL) followed by addition of 2 µM (BMDMs, peritoneal macrophages, PBMCs), 6 µM (THP-1 differentiated macrophages) nigericin (Sigma-Aldrich, N7143-5MG), 2 mM (peritoneal macrophages) ATP or 50 µM (THP-1 differentiated macrophages) R837 (InvivoGen, tlrl-imqs) for 30–60 min. To activate the AIM2 inflammasome, THP-1 macrophages or peritoneal macrophages were primed with LPS (100 ng/mL) for 3 h followed by (Poly(dA:dT), 2 µg/mL) for 6 h using LyoVecTM (InvivoGen, tlrl-patc) according to the manufacturer’s protocol. To activate the NLRC4 inflammasome, BMDMs were primed with LPS (100 ng/mL) for 3 h followed by (flagellin isolated from P. aeruginosa, 2 or 5 µg/mL) for 3 h using FLA-PA Ultrapure (InvivoGen, tlrl-pafla) according to the manufacturer’s protocol. To activate the non-canonical inflammasome, peritoneal macrophages were stimulated with LPS (100 ng/mL) for 3 h followed by intracellular LPS delivery (1 µg/mL) by transfection using lipofectamine 2000 (Invitrogen, 11668027), according to the manufacturer’s protocol, in OptiMEM (Gibco, 31985-070) for 16 h. HNE (0.3–10 µM) (Millipore Sigma, 393204), alkyne-HNE (1–20 µM) (Cayman Chemical, 13265) or HNE-GSH (3–10 µM) (Cayman Chemical Company, 10627) was added 30 min before the inflammasome activator. Micrographs of cell cultures were obtained under phase contrast illumination using 40X objective (Leica) prior to cell lysis. Multiple random fields were captured for each well. Culture supernatants were collected and centrifuged at 500 × g for 5 min to remove cellular debris. LDH measurement was performed with the CyQUANT™ LDH cytotoxicity assay kit (Thermo Fisher Scientific, C20301) according to the manufacturer’s instructions. Data were plotted normalizing the O.D. value obtained in wells treated with Triton X-100 (0.1%) as 100%. THP-1 macrophages or BMDMs were seeded in 96-well plates (2 × 104 cells/well) one day before the experiments. Cells were washed twice and incubated with LPS (100 ng/mL) in XF based medium (Agilent, 103334-100) supplemented with 4.5 g/L glucose, 2 mM glutamine, 1 mM sodium pyruvate, and 1 mM HEPES buffer at final pH7.4 for 3 h. After 3 h LPS stimulation, SYTOX Green (final concentration 1 µM) (Thermo Fisher Scientific, S7020) was added together with nigericin (2 µM for BMDMs; 6 µM for THP-1 macrophages) and fluorescence signals (Excitation wavelength: 485 nm, Emission wavelength: 550 nm) were analyzed using FLUOstar OPTIMA plate reader (BMG Labtech) at 36 oC for 120 min. The percentage cell death was calculated by normalizing fluorescence signals from cells treated with Triton X-100 (0.1%). Nrf2 or NF-κB p65 nuclear translocation was measured by immunofluorescence staining. THP-1 monocytes were seeded on glass coverslips in 12- well plates and differentiated as described above. Cells were treated with LPS (Nrf2 for 3 h; p65 for 1 h) and were washed twice with cold PBS and fixed with 4% formaldehyde in PBS for 10 min. After washing with PBS, the cells were incubated with blocking solution (10% normal goat serum, and 0.1% Triton X-100 in PBS) for 1 h followed by incubating with the Nrf2 (Abcam, ab31163) 1:500; NF-κB p65 (D14E12) (Cell Signaling Technology, 8242) 1:500; antibody overnight. Cells were washed three times with PBS and incubated with the goat anti-rabbit IgG (H + L) secondary antibody, alexa fluor 488 (Thermo Fisher Scientific, A-11034) for 1 h in PBS in the dark. After three washes with PBS, samples were mounted using fluoromount-G-DAPI (SouthernBiotech, 0100-20). Immunofluorescence was analyzed by confocal microscopy. Nuclear translocation was evaluated using NIH ImageJ software with an intensity ratio nuclei cytoplasm macro. Quantification of translocation was expressed as the percentage of intensity in nucleus over total cell [86]. The data points shown are from 3 separate experiments with duplicate or triplicate wells. Around 200 cells from at least 6 images were quantified for each experimental group. Plasma, BAL fluid and peritoneal fluid levels of IL-1 β (Invitrogen 88-7013-22), IL-18 (Invitrogen, BMS618-3) and TNF-α (BioLegend, 430904) collected from mouse studies were determined by ELISA kits according to the manufacturer’s instructions. For in vitro studies, culture media was collected immediately after treatment. Samples were cleared by centrifugation at 16,000 ×g for 5 min and stored at −20 °C. HNE (MyBioSource.com, MBS7606509), Human and mouse IL-1 β (BioLegend, 437004 and 432604) were measured in culture supernatants by ELISA. RNA was extracted from lung tissue or cultured cells using RNeasy kit (Qiagen, 74106) according to the manufacturer’s instructions. Complementary DNA was synthesized from 0.5 μg RNA by iScript™ cDNA Synthesis Kit (Bio-Rad, 1708891). Amplification reactions contained a target specific fraction of cDNA and 1 μM forward and reverse primers in iQ™ SYBR® Green Supermix (Bio-Rad, 1708882). Fluorescence was monitored and analyzed in a CFX connect real-time PCR system (Bio-Rad). Gene expression was normalized to β-actin using the delta delta cycle threshold method. Amplification of specific transcripts was confirmed by melting curve analysis at the end of each PCR experiment. The primers used are as follows: Mouse GCLC (Forward: AGATGATAGAACACGGGAGGAG, Reverse: TGATCCTAAAGCGATTGTTCTTC); Mouse Ferroportin-1(Forward: ACCCATCCCCATAGTCTCTGT, Reverse: ACCGTCAAATCAAAGGACCA); Mouse β-actin (Forward: TTCAACACCCCAGCCATGT, Reverse: GTAGATGGGCACAGTGTGGGT); Mouse IL-1β(Forward: GAGTGTGGATCCCAAGCAAT, Reverse: ACGGATTCCATGGTGAAGTC); Mouse IL-6 (Forward: GAGGATACCACTCCCAACAGACC, Reverse: AAGTGCATCATCGTTGTTCATACA); Mouse TNF-α (Forward: TCTTCTCATTCCTGCTTGTGG, Reverse: GGTCTGGGCCATAGAACTGA); Mouse NLRP3 (Forward: TTCCCAGACACTCATGTTGC, Reverse: AGAAGAGACCACGGCAGAAG); SAA3 (Forward: TTTCTCTTCCTGTTGTTCCCAGTC, Reverse: TCACAAGTATTTATTCAGCACATTGGGA). Proteins were separated by SDS-PAGE through 10% acrylamide gels and transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in Tween-TBS and reacted with the indicated antibody: NLRP3 (AdipoGen, AG-20B-0014-C100) 1:1000; Caspase-1(AdipoGen, AG-20B-0042-C100) 1:1000; GSDMD (Abcam, ab209845) 1:1000; IL-1β (GeneTex, GTX10750) 1:2000; ASC (Santa Cruz Biotechnology, sc-514414) 1:1000; GAPDH (Millipore, MAB374) 1:2000 β-actin (Cell Signaling Technology, 4970) 1:4000; Phospho-NF-κB p65 (Ser536) (93H1) (Cell Signaling Technology, 3033 S) 1:1000; IκB-α (L35A5) (Cell Signaling Technology, 4814) 1:1000; Acetyl-α-Tubulin (Lys40) (5335 T) 1:1000; NEK7 (Abcam, ab133514) 1:1000 overnight. Membranes were rinsed and incubated with horseradish peroxidase conjugated secondary antibody (Anti-mouse IgG, HRP-linked Antibody (Cell Signaling Technology, 7076), Anti-rabbit IgG, HRP-linked Antibody (Cell Signaling Technology, 7074), Anti-goat IgG, HRP-linked Antibody (Jackson Immuno Research, 805-035-180). Reactive proteins were detected by enhanced chemiluminescence, visualized by exposure to radiographic film and quantified by scanning densitometry normalized to β-actin expression measured in each sample on the same gel. All of the full and uncropped western blots were listed in supplemental material. ASC-GFP-expressing THP-1differentiated macrophages were seeded on glass coverslips in 12- well plates. Cells were incubated with 100 ng/ml LPS for 3 h followed by incubation with 6 µM nigericin for 2 h. Cells were fixed in 4% paraformaldehyde for 10 min. After three washes with 1x PBS, cells were mounted using fluoromount-G-DAPI. Immunofluorescence was analyzed by confocal microscope. The GFP 488 nm fluorescent signals were acquired by confocal microscopy. The data points represent biological replicates from 3 separate experiments. Around 200 cells from at least 6 images were quantified for each experimental group. For ASC oligomer cross-linking, cells were lysed in buffer (0.5% Triton × 100, 20 mM HEPES-KOH, pH 7.5, 150 mM KCl, and complete protease and phosphatase inhibitor cocktail) on ice by syringing 10 times through a G26 needle. The cell lysates were centrifuged at 6000 rpm at 4 °C for 10 min. The pellets were resuspended in PBS and crosslinked with 2 mM disuccinimidyl suberate (Thermo Fisher Scientific, 21655). The cross-linked pellets were centrifuged at 15000 rpm for 15 min and dissolved directly in a 1x SDS sample buffer. THP-1differentiated macrophages were seeded on glass coverslips in 12- well plates. Cells were incubated with 100 ng/ml LPS for 3 h followed by incubation with 2 µM Mitosox red (Thermo Fisher Scientific, M36008) for 30 min. Cells were washed twice with medium and stimulated with nigericin for 1 h. Cells were fixed in 4% paraformaldehyde for 10 min. After three washes with 1x PBS, cells were mounted using fluoromount-G-DAPI. Immunofluorescence was analyzed by confocal microscope. The 510/580 nm fluorescent signals were acquired by confocal microscopy. Cells were lysed in IP Lysis buffer (20 mM HEPES-KOH, 150 mM NaCl, and 0.5% v/v Triton-100x) containing protease inhibitor cocktail, and treated with 10 mM sodium borohydride (Sigma-Aldrich, 452882) for 30 min. Click-chemistry was performed with Molecular Probes Click iT Protein Reaction Buffer Kit (Thermo Fisher Scientific, C10276) according to the manufacturer’s instructions. Briefly, The Cu(I)-catalyzed click reaction was initiated by adding Click-iT reaction buffer and 50 µl of Azide-PEG3-biotin in 50 mM (Cayman Chemical, 23419) to each sample and incubated on a rotator for 20 min at room temperature. Protein was precipitated and the pellet was washed once with 100 µl of ice-cold methanol and centrifuged at 15,000 x g for 5 min. Methanol was removed and the pellets were re-suspended in 50 µl IP Lysis buffer containing protease inhibitor cocktail. Protein concentrations were estimated assuming no protein loss occurred during the click and precipitation procedure. To perform biotin affinity precipitation, clicked lysates were loaded with 20 µl of Dynabeads™ MyOne™ Streptavidin C1 (Invitrogen, 65001). The Dynabeads were equilibrated to buffer by three 100 µl washes. Thirty micrograms of “clicked” lysates were loaded onto the equilibrated beads. The volume was brought up to 100 µl with IP lysis buffer, and lysates were incubated with beads for 1 h at room temperature on a rotator. After incubation, the beads were washed six times with 100 µl volumes of IP lysis buffer. The bound proteins were eluted using 100 µl of 1X sample lysis buffer, by vortexing and heating at 95 °C for 10 min. The supernatant was collected and stored at −20 °C for western blot analysis. Cells were lysed in IP buffer on ice by syringing 10 times through a G26 needle. The supernatant was collected as the whole cell lysate (WCL) sample. One mg total protein from WCL was incubated with anti-NEK7 antibody (Abcam, ab133514), anti-NLRP3 antibody (AdipoGen, AG-20B-0014-C100), ChromPure Rabbit IgG (Jackson Immuno Research, 011-000-003), or ChromPure Mouse IgG (Jackson Immuno Research, 015-000-00) overnight. Agarose beads (Roche, C755B62) was added to the remaining supernatant, which was subsequently incubated at 4 oC in a ferris wheel mixer for 3 h. IP samples were subsequently centrifuged at 200 x g for 2 min at 4 oC, supernatant removed, and beads washed five times with 1 mL low stringency lysis buffer. The immune complexes were eluted by addition of 100 µL of 1X sample lysis buffer, boiled for 5 min and analyzed by western blot. Unless otherwise noted, in vitro experiments were repeated as three independent procedures, with duplicate or triplicate wells averaged prior to statistical analysis. All data were presented as mean ± SD. GraphPad Prism 8.0 was used for statistical analysis. Comparisons between two groups after stimulation were analyzed by two-way ANOVA. HNE dose response experiments in cell cultures were analyzed by one-way ANOVA followed by post hoc T tests using Bonferroni correction for multiple comparisons. P values were indicated as follow: * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001.THP-1 macrophage differentiation
ASC-GFP-overexpressed THP-1 macrophage
Overexpressed GSDMD-NT HEK293T cells
HEK293As overexpressed with NLRP3, NEK7, and ASC-GFP
LPS stimulation
Inflammasome stimulation
Cell morphology
Cell death LDH assay
Cell death by SYTOXTM green
Nuclear translocation of Nrf2 and p65
Cytokine and HNE assays
RNA extraction and Real-time PCR
Western blot
ASC speck formation
ASC oligomerization
MitoSOX red imaging
Click chemistry and biotin affinity precipitation
Co-immunoprecipitation
Statistics
Data availability
All data generated or analysed during this study are included in this published article and in its supplementary file.
References
McKenzie BA, Dixit VM, Power C. Fiery cell death: Pyroptosis in the central nervous system. Trends Neurosci. 2020;43:55–73.
Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature 2000;406:782–7.
Ulevitch RJ, Tobias PS. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr Opin Immunol. 1999;11:19–22.
Samir P, Kesavardhana S, Patmore DM, Gingras S, Malireddi RKS, Karki R, et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 2019;573:590–4.
Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 2010;464:1357–61.
Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008;320:674–7.
Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21.
Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature 2012;481:278–86.
Guo H, Callaway JB, Ting JP. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–87.
He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25:1285–98.
Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015;526:666–71.
Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015;526:660–5.
Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl J Med. 2017;377:1119–31.
Liang JJ, Fraser IDC, Bryant CE. Lipid regulation of NLRP3 inflammasome activity through organelle stress. Trends Immunol. 2021;42:807–23.
Gil L, Siems W, Mazurek B, Gross J, Schroeder P, Voss P, et al. Age-associated analysis of oxidative stress parameters in human plasma and erythrocytes. Free Radic Res. 2006;40:495–505.
Siems WG, Grune T, Esterbauer H. 4-Hydroxynonenal formation during ischemia and reperfusion of rat small intestine. Life Sci. 1995;57:785–9.
Schaur RJ, Siems W, Bresgen N, Eckl PM. 4-Hydroxy-nonenal-A Bioactive Lipid Peroxidation Product. Biomolecules 2015;5:2247–337.
Awasthi YC, Ramana KV, Chaudhary P, Srivastava SK, Awasthi S. Regulatory roles of glutathione-S-transferases and 4-hydroxynonenal in stress-mediated signaling and toxicity. Free Radic Biol Med. 2017;111:235–43.
Dalleau S, Baradat M, Gueraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–30.
Jaganjac M, Milkovic L, Gegotek A, Cindric M, Zarkovic K, Skrzydlewska E, et al. The relevance of pathophysiological alterations in redox signaling of 4-hydroxynonenal for pharmacological therapies of major stress-associated diseases. Free Radic Biol Med. 2020;157:128–53.
Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–44.
Van Hall-Beauvais A, Zhao Y, Urul DA, Long MJC, Aye Y. Single-Protein-Specific Redox Targeting in Live Mammalian Cells and C. elegans. Curr Protoc Chem Biol. 2018;10:e43.
Abulafia DP, de Rivero Vaccari JP, Lozano JD, Lotocki G, Keane RW, Dietrich WD. Inhibition of the inflammasome complex reduces the inflammatory response after thromboembolic stroke in mice. J Cereb Blood Flow Metab. 2009;29:534–44.
Siow RC, Ishii T, Mann GE. Modulation of antioxidant gene expression by 4-hydroxynonenal: Atheroprotective role of the Nrf2/ARE transcription pathway. Redox Rep. 2007;12:11–5.
Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, et al. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: Activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res. 2004;94:609–16.
Page S, Fischer C, Baumgartner B, Haas M, Kreusel U, Loidl G, et al. 4-Hydroxynonenal prevents NF-kappaB activation and tumor necrosis factor expression by inhibiting IkappaB phosphorylation and subsequent proteolysis. J Biol Chem. 1999;274:11611–8.
Zhang Y, Sano M, Shinmura K, Tamaki K, Katsumata Y, Matsuhashi T, et al. 4-hydroxy-2-nonenal protects against cardiac ischemia-reperfusion injury via the Nrf2-dependent pathway. J Mol Cell Cardiol. 2010;49:576–86.
Timucin AC, Basaga H. Pro-apoptotic effects of lipid oxidation products: HNE at the crossroads of NF-kappaB pathway and anti-apoptotic Bcl-2. Free Radic Biol Med. 2017;111:209–18.
Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006;440:228–32.
Hennig P, Garstkiewicz M, Grossi S, Di Filippo M, French LE, Beer HD. The Crosstalk between Nrf2 and Inflammasomes. Int J Mol Sci. 2018;19:562.
Garstkiewicz M, Strittmatter GE, Grossi S, Sand J, Fenini G, Werner S, et al. Opposing effects of Nrf2 and Nrf2-activating compounds on the NLRP3 inflammasome independent of Nrf2-mediated gene expression. Eur J Immunol. 2017;47:806–17.
Zhao C, Gillette DD, Li X, Zhang Z, Wen H. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J Biol Chem. 2014;289:17020–9.
Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–57.
Song X, Long D. Nrf2 and Ferroptosis: A new research direction for neurodegenerative diseases. Front Neurosci. 2020;14:267.
Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem Biol. 2016;11:3214–25.
Jo EK, Kim JK, Shin DM, Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol Immunol. 2016;13:148–59.
Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–60.
Stutz A, Horvath GL, Monks BG, Latz E. ASC speck formation as a readout for inflammasome activation. Methods Mol Biol. 2013;1040:91–101.
Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21.
Carpe N, Mandeville I, Kho AT, Qiu W, Martin JG, Tantisira KG, et al. Maternal allergen exposure reprograms the developmental lung transcriptome in atopic and normoresponsive rat pups. Am J Physiol Lung Cell Mol Physiol. 2012;303:L899–911.
Cheng KT, **ong S, Ye Z, Hong Z, Di A, Tsang KM, et al. Caspase-11-mediated endothelial pyroptosis underlies endotoxemia-induced lung injury. J Clin Invest. 2017;127:4124–35.
Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016;535:153–8.
Hu JJ, Liu X, **a S, Zhang Z, Zhang Y, Zhao J, et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat Immunol. 2020;21:736–45.
He Y, Zeng MY, Yang D, Motro B, Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016;530:354–7.
Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, et al. A genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) screen Identifies NEK7 as an essential component of NLRP3 inflammasome activation. J Biol Chem. 2016;291:103–9.
Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 2019;570:338–43.
Shi H, Wang Y, Li X, Zhan X, Tang M, Fina M, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–8.
Parker CG, Pratt MR. Click chemistry in proteomic investigations. Cell 2020;180:605–32.
Wang S, Zhao J, Wang H, Liang Y, Yang N, Huang Y. Blockage of P2X7 attenuates acute lung injury in mice by inhibiting NLRP3 inflammasome. Int Immunopharmacol. 2015;27:38–45.
dos Santos G, Rogel MR, Baker MA, Troken JR, Urich D, Morales-Nebreda L, et al. Vimentin regulates activation of the NLRP3 inflammasome. Nat Commun. 2015;6:6574.
Hsu CG, Fazal F, Rahman A, Berk BC, Yan C. Phosphodiesterase 10A is a key mediator of lung inflammation. J Immunol. 2021;206:3010–20.
Danielski LG, Giustina AD, Bonfante S, Barichello T, Petronilho F. The NLRP3 inflammasome and its role in sepsis development. Inflammation 2020;43:24–31.
Griffiths RJ, Stam EJ, Downs JT, Otterness IG. ATP induces the release of IL-1 from LPS-primed cells in vivo. J Immunol. 1995;154:2821–8.
Csala M, Kardon T, Legeza B, Lizak B, Mandl J, Margittai E, et al. On the role of 4-hydroxynonenal in health and disease. Biochim Biophys Acta. 2015;1852:826–38.
Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289–303.
Jia M, Qin D, Zhao C, Chai L, Yu Z, Wang W, et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat Immunol. 2020;21:727–35.
Brown CW, Chhoy P, Mukhopadhyay D, Karner ER, Mercurio AM. Targeting prominin2 transcription to overcome ferroptosis resistance in cancer. EMBO Mol Med. 2021:e13792:1–14.
Shintoku R, Takigawa Y, Yamada K, Kubota C, Yoshimoto Y, Takeuchi T, et al. Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci. 2017;108:2187–94.
Schroder K, Tschopp J. The inflammasomes. Cell 2010;140:821–32.
Kauppinen A, Niskanen H, Suuronen T, Kinnunen K, Salminen A, Kaarniranta K. Oxidative stress activates NLRP3 inflammasomes in ARPE-19 cells-implications for age-related macular degeneration (AMD). Immunol Lett. 2012;147:29–33.
** X, Wang C, Wu W, Liu T, Ji B, Zhou F. Cyanidin-3-glucoside Alleviates 4-Hydroxyhexenal-Induced NLRP3 Inflammasome Activation via JNK-c-Jun/AP-1 Pathway in Human Retinal Pigment Epithelial Cells. J Immunol Res. 2018;2018:5604610.
Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5.
Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: A sensor for metabolic danger? Science 2010;327:296–300.
Evavold CL, Hafner-Bratkovic I, Devant P, D’Andrea JM, Ngwa EM, Borsic E, et al. Control of gasdermin D oligomerization and pyroptosis by the Ragulator-Rag-mTORC1 pathway. Cell. 2021;184:4495–11.
Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018;556:113–7.
Ghosh S, Choudhury S, Chowdhury O, Mukherjee S, Das A, Sain A, et al. Inflammation-induced behavioral changes is driven by alterations in Nrf2-dependent apoptosis and autophagy in mouse hippocampus: Role of fluoxetine. Cell Signal. 2020;68:109521.
Dong Z, Shang H, Chen YQ, Pan LL, Bhatia M, Sun J. Sulforaphane protects pancreatic acinar cell injury by modulating Nrf2-mediated oxidative stress and NLRP3 inflammatory pathway. Oxid Med Cell Longev. 2016;2016:7864150.
Liu X, Zhang X, Ding Y, Zhou W, Tao L, Lu P, et al. Nuclear factor E2-related factor-2 negatively regulates NLRP3 inflammasome activity by inhibiting reactive oxygen species-induced NLRP3 priming. Antioxid Redox Signal. 2017;26:28–43.
Liu X, Zhou W, Zhang X, Lu P, Du Q, Tao L, et al. Dimethyl fumarate ameliorates dextran sulfate sodium-induced murine experimental colitis by activating Nrf2 and suppressing NLRP3 inflammasome activation. Biochem Pharm. 2016;112:37–49.
Polster BM, Nicholls DG, Ge SX, Roelofs BA. Use of potentiometric fluorophores in the measurement of mitochondrial reactive oxygen species. Methods Enzymol. 2014;547:225–50.
Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–15.
Jiang Y, Wang M, Huang K, Zhang Z, Shao N, Zhang Y, et al. Oxidized low-density lipoprotein induces secretion of interleukin-1beta by macrophages via reactive oxygen species-dependent NLRP3 inflammasome activation. Biochem Biophys Res Commun. 2012;425:121–6.
Alberts BM, Bruce C, Basnayake K, Ghezzi P, Davies KA, Mullen LM. Secretion of IL-1beta from monocytes in gout is redox independent. Front Immunol. 2019;10:70.
Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol. 2011;187:613–7.
Marantos C, Mukaro V, Ferrante J, Hii C, Ferrante A. Inhibition of the lipopolysaccharide-induced stimulation of the members of the MAPK family in human monocytes/macrophages by 4-hydroxynonenal, a product of oxidized omega-6 fatty acids. Am J Pathol. 2008;173:1057–66.
Bezbradica JS, Coll RC, Schroder K. Sterile signals generate weaker and delayed macrophage NLRP3 inflammasome responses relative to microbial signals. Cell Mol Immunol. 2017;14:118–26.
Schroder K, Sagulenko V, Zamoshnikova A, Richards AA, Cridland JA, Irvine KM, et al. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 2012;217:1325–9.
McKee CM, Coll RC. NLRP3 inflammasome priming: A riddle wrapped in a mystery inside an enigma. J Leukoc Biol. 2020;108:937–52.
Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73.
Mortimer L, Moreau F, MacDonald JA, Chadee K. NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat Immunol. 2016;17:1176–86.
Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–55.
Kang R, Zeng L, Zhu S, **e Y, Liu J, Wen Q, et al. Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe. 2018;24:97–108 e4.
Mukhopadhyay S, Pluddemann A, Hoe JC, Williams KJ, Varin A, Makepeace K, et al. Immune inhibitory ligand CD200 induction by TLRs and NLRs limits macrophage activation to protect the host from meningococcal septicemia. Cell Host Microbe. 2010;8:236–47.
Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Visualized Exp.: JoVE. 2013;76:50323.
Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl. 1968;97:77–89.
Zhang C, Hsu CG, Mohan A, Shi H, Li D, Yan C. Vinpocetine protects against the development of experimental abdominal aortic aneurysms. Clin Sci (Lond). 2020;134:2959–76.
Acknowledgements
This work was financially supported by National Institute of Health HL134910 (to BCB and CY), HL140958 (to BCB), HL154318 (to CY), Department of Defense DM190884 (to BCB), New York State Department of Health C34726GG (to BCB and CGH), and University of Rochester Environmental Health Sciences Center P30 ES001247 (to BCB).
Author information
Authors and Affiliations
Contributions
CGH, MS, CY, BCB designed research; CGH, CLC, CZ, MS performed research; BCB contributed new reagents/ analytic tools; CGH, CZ analyzed data; CGH, CY, BCB wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Ethics statement
All of the experiments were approved by the University Committee on Animal Use For Research (UCAR) at the University of Rochester and followed National Institutes of Health guidelines for experimental procedures on mice. Human blood samples from healthy donors were collected and processed at the University of Rochester Medical Center following Institutional Review Board approval.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Oberst
Rights and permissions
About this article
Cite this article
Hsu, C.G., Chávez, C.L., Zhang, C. et al. The lipid peroxidation product 4-hydroxynonenal inhibits NLRP3 inflammasome activation and macrophage pyroptosis. Cell Death Differ 29, 1790–1803 (2022). https://doi.org/10.1038/s41418-022-00966-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00966-5
- Springer Nature Limited
This article is cited by
-
Signaling pathways and potential therapeutic targets in acute respiratory distress syndrome (ARDS)
Respiratory Research (2024)
-
Bhlhe40 deficiency attenuates LPS-induced acute lung injury through preventing macrophage pyroptosis
Respiratory Research (2024)
-
The role of inflammasomes in human diseases and their potential as therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
Macrophage polarization and metabolism in atherosclerosis
Cell Death & Disease (2023)
-
Pnpt1 mediates NLRP3 inflammasome activation by MAVS and metabolic reprogramming in macrophages
Cellular & Molecular Immunology (2023)