Abstract
Objective
Metabolic disturbance of lysophosphatidylcholine (LPC) is related with dyslipidemia. Therefore, eight single-nucleotide polymorphisms (SNPs) were selected from LPC metabolic enzymes to study their associations with obesity and serum levels of lipids.
Methods
A total of 3305 children were recruited from four independent studies. Eight SNPs of LPC metabolic enzymes were selected and genotyped with the matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS). The multivariable linear regression model was applied to detect the associations of eight SNPs with obesity-related phenotypes and levels of lipids in each study. Meta-analyses were used to combine the results of four studies.
Results
Only SNP rs4420638 of APOC-1 gene was associated with serum lipids even after Bonferroni correction. The rs4420638 was positively associated with TC (β = 0.15, P = 8.59 × 10−9) and low-density-lipoprotein-cholesterol (LDL-C, β = 0.16, P = 9.98 × 10−14) individually.
Conclusion
The study firstly revealed the association between APOC-1/rs4420638 and levels of serum lipids in Chinese children, providing evidence for susceptible gene variants of dyslipidemia.
Similar content being viewed by others
Introduction
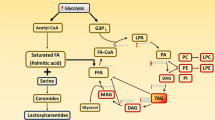
Lysophosphatidylcholine (LPC) has been gradually considered as a key biomarker involved in the pathogenesis of cardiovascular disease1 and obesity.2 LPC derived from cleaving of phosphatidylcholine (PC) under the action of phospholipase A2 (PLA2) and/or by transfer of fatty acids to free cholesterol with the help of lecithin-cholesterol acyltransferase.3,4 In turn, LPC and PLA2 can be converted back to PC via the enzyme lysophosphatidylcholine acyltransferase (LPCAT, encoded by gene LPCAT1) in the presence of Acyl-CoA,5 and this process is named Lands cycle. Overproduction of LPC could result in elevated activity of lipoprotein-associated phospholipase A2 (Lp-PLA2) in the circulation and the Lp-PLA2 was encoded by the gene named PLA2G7.6 Lp-PLA2 has much higher affinity bounding to low-density-lipoprotein (LDL) than high-density-lipoprotein (HDL). Previous research indicated that participants with high LDL levels also had high Lp-PLA2 activity. Additionally, certain number of single-nucleotide polymorphisms (SNPs) of PLA2G7 were associated with activity of Lp-PLA2.7 Secretory phospholipase A2 group IIA (PLA2G2A) is a member of secretory phospholipase A2 (sPLA2), a subgroup from PLA2, which has been involved in the regulation of cholesterol and HDL levels in published research.8 Several studies have investigated the associations between genetic variants of LPC metabolic enzymes, such as rs1805017, rs1421378, and rs2216465 of PLA2G7;7 rs1774131 and rs3753827 of PLA2G2A;9 rs7737692 of LPCAT1 (ref. 10) and the activity of each corresponding protein. The rs4420638 located at 340 bp from the 3′ end of the apolipoprotein C-I gene (APOC-1) was significantly related to the activity of Lp-PLA2 both in Caucasia7 and Chinese population.11 Furthermore, rs4420638 had significant association with total cholesterol (TC) and HDL levels in one study among German children.12 Nonetheless, no study investigated the associations of these genetic variants with obesity and serum lipids levels in Chinese children. Therefore, we conducted the present study to discover the associations of the genetic variants of metabolic enzymes of LPC with obesity-related phenotypes and serum lipids in Chinese children.
Methods
Study population
All the participants were aged between 6 and 18 years old and recruited from four independent studies which were carried out in Bei**g, China. The studies were Adolescent Lipids, Insulin Resistance and candidate genes (ALIR carried out in 2004 and included 937 students),13 Comprehensive Prevention project for Overweight and Obese Adolescent (CPOOA carried out during 2006-2008 and included 1093 students),14 School-Based Comprehensive Intervention on Childhood Obesity (SCICO conducted during 2009–2010 and contained 819 students),15 and School-Based Physical Activity Intervention on Childhood Obesity (SPAICO conducted and completed in 2012 and contained 456 students),16 respectively. All the detailed information of four studies can be referred from previous publications.13,14,15,16 Finally, 3305 unrelated Chinese Han Children were included in the study, including 1262 normal weight children, 1014 overweight children, and 1029 obesity children. The exclusion and inclusion criteria for all four studies were consistent. Children who had heart, liver, lung, or kidney diseases or endocrine disorder diseases were excluded. The ALIR, CPOOA, and SPAICO studies were approved by the Ethics Committee Board of Peking University Health Science Center and the SCICO study were approved by the Ethics Committee Board of Chinese Center for Disease Control and Prevention. Additionally, all the participants and their parents had the written informed consents.
Anthropometric and lipids measurements
Height, weight, waist, and hip circumferences were measured with consistent methods based on the standard protocols across four studies.13,14 Body mass index (BMI) was calculated by weight (kg) divided by height (m2). Waist to hip ratio (WHR) was calculated by waist circumference (cm) divided by hip circumference (cm) and waist to height ratio (WHtR) was calculated by waist circumference(cm) divided by height (cm). For children aged 7–18 years, we used uniform BMI percentile criteria, which were determined in a representative Chinese population.17 Children with an age- and sex-specific BMI < 15th were classified as underweight and were excluded. Then, children with an age- and sex-specific BMI ≥ 95th percentile were defined as obese and those with an age- and sex-specific 95th > BMI ≥ 85th percentile were defined as overweight. Lastly, the rest children were classified as normal weight. For children who were 6 years old, the Chinese national screening criteria for overweight and obesity among school-age children and adolescents were used.18
The fasting venous blood from each participant was collected in the morning. Genomic DNA were extracted by using the phenol–chloroform extraction method (ALIR, CPOOA, and SCICO) or salt extraction method (SPAICO). TC, triglyceride (TG), LDL-cholesterol (LDL-C), and HDL-cholesterol (HDL-C) were detected with the biochemical autoanalyzer (ALIR and CPOOA: Hitachi7060, Tokyo, Japan; SCICO and SPAICO: Olympus AU400, Tokyo, Japan). All the detections were performed according to the manufacturer’s instructions.
Genetic variants selection and genoty**
The SNPs of LPC metabolic enzymes were selected based on previous published literatures of PLA2G7, APOC-1, PLA2G2A, and LPCAT1. Further, power calculation was performed with Quanto software (University of Southern California, Los Angeles, CA). According to the previous literatures, the effect size of genetic variants could range from 0.01 to 0.10,19 we select 0.05 as the effect size to conduct the power calculation. Under the additive model, with the assumed effect size (β = 0.05), the mean value 4.0, and standard deviation 0.5 of TG, we found when effect allele frequency ≥0.12 the statistical power to detect a positive association would be ≥75% with our sample size. Additionally, Hardy–Weinberg equilibrium (HWE) tests in normal weight had P > 0.05. Finally, eight SNPs from four genes (PLA2G7, APOC-1, PLA2G2A, and LPCAT1) were selected (Table S1).
Genoty** of eight SNPs were carried out with matrix-assisted laser desorption ionization time of flight mass spectrometry (the MassArray system, Agena Bioscience Inc., San Diego, CA, US). All the call rates were above 98.9%. Genoty** was performed with 0.5% randomly selected duplicated samples and the consistent rates were 100%. More detailed procedure could be referred from previous studies.20
Bioinformatic analyses
Expression quantitative trait loci (eQTL) were checked with Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/home/) and certain tissues were selected to present normalized effect size and P value (Table S2).
Statistical analyses
The differences of basic characteristics among normal weight, overweight, and obesity children were compared with one-way analysis of variance (ANOVA) for continuous variables and χ2 test for categorical variables, respectively. Multivariable linear regressions were applied to investigate the association between genetic variants and obesity-related phenotypes adjusted for sex, age, age squared, and for the associations between genetic variants and serum lipids adjusted for BMI additionally. Both regression analyses were under the additive model. Furthermore, meta-analyses were carried out to pool the results of four studies. Heterogeneity of four studies was detected with inconsistency index (I2). The random-effect model was applied when I2 > 0.5 or P < 0.1, otherwise, the fix-effect model was adopted. To be noticed, lack of the measurement of hip circumference in SCICO study, the meta-analyses of associations between genetic variants and hip circumference, WHR were performed with the other three studies. Bonferroni correction was carried out, with two-sided α < 6.94 × 10−4 [0.05/(8 SNPs × 9 phenotypes)]. All statistical analyses were conducted with SPSS 16.0 (IBM Corp., Armonk, NY, US). Meta-analyses were performed with Stata 14.0 (Stata Corp., College Station, TX, US)
Results
The characteristics of study subjects are shown in Table 1. All the variables were significantly different across three groups. Particularly, the obesity group had highest proportion of boys (67.2%). Consistently, the concentrations of TC, TG, and LDL-C steadily increased with BMI categories (normal weight < overweight < obesity), while HDL-C decreased (normal weight > overweight > obesity).
We analyzed the associations between these SNPs and obesity related phenotypes (Table 2). Results indicated that no significant association of these SNPs with BMI, waist circumference, hip circumference, WHR, or WHtR. Only rs1805017 of PLA2G7 had moderate association with WHR (β = 0.005, P = 0.03); however, after Bonferroni correction, the association did not exist.
The associations between these SNPs and levels of serum lipids were further analyzed (Table 3). Two SNPs of rs1805017 and rs4654990 had statistically significant associations with TG (β = 0.04, P = 0.001) and TC (β = 0.05, P = 0.01) individually. However, those associations did not exist after Bonferroni correction. While rs4420638 had significant associations with TC (β = 0.15, P = 8.59 × 10−9), TG (β = 0.10, P = 0.01) and LDL-C (β = 0.16, P = 9.98 × 10−14), which remained significant for TC and TG after Bonferroni correction.
The SNP of rs4420638 was further selected to explore the expression of their coding genes and the eQTL in GETx (https://gtexportal.org/home/). The priority of selection was tissue (adipose-subcutaneous) and the P value of the eQTL (Table S2). The SNP of rs4420638 mainly cis-regulated the expression of APOC-1 and APOE.
Discussion
In the present study, association analyses between several SNPs of LPC metabolic enzymes and obesity-related phenotypes or serum lipids were carried out. The results revealed that the SNP rs4420638 was associated with serum lipids even after Bonferroni correction in Chinese children.
Lp-PLA2 is a member of PLA2 and also named as platelet- activating factor acectylhydrolase (PAF-AH), which is produced by hematopoietic cells and highly expressed in the spleen.21 Additionally, the HDL and LDL had different binding site on the Lp-PLA2.21 The SNP rs1805017 is a non-synonymous variation located in the PLA2G7 and encoded the R92H in the protein of Lp-PLA2. Previous Caucasian population studies indicated that rs1805017 T allele had positive association with Lp-PLA2 mass. Furthermore, controlling the confounding of mass, rs1805017 had significant association with the activity of Lp-PLA2 as well.7 However, whether the SNPs of Lp-PLA2 had associations with serum lipids were explored by a few studies. Suchindran et al. found no association between serum lipids and PLA2G7 genotypes (including rs1805017), but Zhou et al.22 found rs76863441 but not rs1805017 of PLA2G7 had association with higher HDL and TG in preeclampsia women. In the present study, we detected nominal associations of rs1805017 with TG and WHR and no association with LDL/HDL. However, no statistical association remained after Bonferroni corrections. One study carried out with rare mutations did not found association with BMI and coronary heart disease (CHD), which was consistent with our present study.23 The plausible reason might be that the frequencies of those mutations are generally low in all populations; it is hard to detect an association.
The SNP rs4420638 is located near the APOE–APOC1–APOC4–APOC2 cluster and it had strong association with the activity of Lp-PLA2, LDL-C, and HDL-C in previous studies performed in Caucasian population.7,24,25 One GWAS study illustrated that rs4420638 associated with the activity of Lp-PLA2 even after adjusting with mass of Lp-PLA2. Additionally, one study carried out in 4192 Chinese Han population revealed that rs4420638 was associated with TC, LDL-C, and TC/HDL-C ratio,26 providing evidence in multi-ethnic populations (Caucasian, East Asian, South Asian, and African American). Those results were mainly in line with our present study. APOC1 protein was found to be highly expressed in the liver and adrenal gland (Table S2). The G allele reduced the expression of APOC1 both in adrenal gland and skin according to the GTEx profiles, with each copy of G allele associated with increasing Lp-PLA2 activity.7 Consequentially, higher Lp-pLA2 activity was connected with higher concentration of LDL-C,27 particularly small LDL28 and electronegative LDL.29 Most researches revealed the association between rs4420638 and CHD, which indicated that rs4420638 could be a risk factor for predicting atherosclerosis and related vascular diseases. Based on the significant function of rs4420638 in dyslipidemia and CHD, molecular mechanisms of genetic variants in combination with Lp-PLA2 inhibitor should be studied in vivo and in vitro to confirm the results generated from association studies in populations.
PLA2G2A also known as sPLA2IIa is a member of secretory phospholipase A2 group which is highly expressed in the coronary and subcutaneous adipose. It had low molecular mass which can be secreted into the extra cellular and was first detected from the platelets and synovial fluids of arthritis patients.30,31 PLA2G2A hydrolyze LDL and produce small-dense LDL particles.32 In the case of acute phase HDL was a better substrate of PLA2G2A.32 In the present study, rs4654990 had nominal association with serum lipds; however, after Bonferroni correction, the statistical significance varnished. Some researches have indicated that PLA2G2A was involved in insulin metabolism and CVD.8,33,34 However, only one genetic variation study which was carried out with 1869 sudden cardiac arrest (SCA) patients implied that rs4654990 G allele carriers had higher risk of SCA with circulating n−3 and trans fatty acids.35 No association study with lipoproteins was conducted in Asia population yet. From the GTEx portal, we observed that rs4654990 G allele had cis regulation of OTUD3 in adrenal gland, thyroid, and subcutaneous adipose and the effect allele G could increase the expression of OTUD3 (Table S2). OTUD3 is a deubiquitinating enzyme that might stabilize the protein of interest, trigger accumulation of vicious proteins, and have impact on the lipoproteins.
Till now, only two studies investigated the SNP rs7737692. One was a case–control study that found rs7737692 lower the risk of SCA in the discovery and replication phase.10 Another research found rs7737692 interacted with red and processed meat intake which could increase the risk of colorectal cancer.36 The SNP rs7737692 is located on the downstream 500 bases of LPCAT1. The previous research indicated that LPCAT1 was involved in the fatty acid and LPC metabolism.37 However, in the present study, we did not detect its association with obesity or lipids. The plausible reason might be the age of our study population was much younger than the previous research.
The strength of present study is that it was the first to discover the associations of genetic variants of LPC metabolic enzymes with obesity-related phenotypes and serum lipids in Chinese children. Moreover, all the participants were children without chronic disease confounding. Additionally, we further confirmed that some genetic variants which influenced serum lipids may not be associated with obesity. The limitation of this study should be noticed as well, for the selection of genetic variants was based on the sample size and statistical power so that a certain number of rare genetic variants was not studied.
In conclusion, the present study identified the associations of rs1805017 (PLA2G7) and rs4420638 (APOC1) with levels of serum lipids. The study results indicated that genetic variants of LPC metabolic enzymes might be associated with risk of dyslipidemia, providing evidence for susceptible gene variants of dyslipidemia and potential precision treatment.
References
Law, S. H. et al. An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. 20, 1149 (2019).
Wang, Y. et al. Lipidomic profile revealed the association of plasma lysophosphatidylcholines with adolescent obesity. Biomed. Res. Int. 2019, 1382418 (2019).
Ridgway, N. & McLeod, R. Biochemistry of Lipids (Elsevier, 2016).
Gauster, M. et al. Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J. Lipid Res. 46, 1517–1525 (2005).
Wang, L. P. et al. Metabolic Interactions between the lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis develo** seeds. Plant Cell 24, 4652–4669 (2012).
Lp-PLA Studies Collaboration et al. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet 375, 1536–1544 (2010).
Suchindran, S. et al. Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham Heart Study. PLoS Genet. 6, e1000928 (2010).
Kuefner, M. S. et al. Secretory phospholipase A2 group IIA modulates insulin sensitivity and metabolism. J. Lipid Res. 58, 1822–1833 (2017).
Wootton, P. T. et al. Tagging-SNP haplotype analysis of the secretory PLA2IIa gene PLA2G2A shows strong association with serum levels of sPLA2IIa: results from the UDACS study. Hum. Mol. Genet. 15, 355–361 (2006).
Lemaitre, R. N. et al. Common variation in fatty acid metabolic genes and risk of incident sudden cardiac arrest. Heart Rhythm 11, 471–477 (2014).
Qi, Y. et al. A previously unreported impact of a PLA2G7 gene polymorphism on the plasma levels of lipoprotein-associated phospholipase A2 activity and mass. Sci. Rep. 6, 37465 (2016).
Breitling, C. et al. Genetic contribution of variants near SORT1 and APOE on LDL cholesterol independent of obesity in children. PLoS ONE 10, e0138064 (2015).
Wang, D. et al. Association of the MC4R V103I polymorphism with obesity: a Chinese case-control study and meta-analysis in 55,195 individuals. Obesity (Silver Spring) 18, 573–579 (2010).
Wang, H. J., Zhang, H., Zhang, S. W., Pan, Y. P. & Ma, J. Association of the common genetic variant upstream of INSIG2 gene with obesity related phenotypes in Chinese children and adolescents. Biomed. Environ. Sci. 21, 528–536 (2008).
Song, Q. Y. et al. Association study of three gene polymorphisms recently identified by a genome-wide association study with obesity-related phenotypes in chinese children. Obes. Facts 10, 179–190 (2017).
Li, X. H. et al. Effectiveness of a school-based physical activity intervention on obesity in school children: a nonrandomized controlled trial. BMC Public Health 14, 1282 (2014).
Ji, C. Y. Working Group on Obesity in C. Report on childhood obesity in China (1)−body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed. Environ. Sci. 18, 390–400 (2005).
National Health Commission of the People’s Repulic of China. Screening for Overweight and Obesity among School-Age Children and Adolescents (National Health Commission of the People’s Repulic of China, Bei**g, 2018).
Varga, T. V. et al. Genetic determinants of long-term changes in blood lipid concentrations: 10-year follow-up of the GLACIER study. PLoS Genet. 10, e1004388 (2014).
Wang, H. J. et al. Association of common variants identified by recent genome-wide association studies with obesity in Chinese children: a case-control study. BMC Med. Genet. 17, 7 (2016).
Stafforini, D. M. Biology of platelet-activating factor acetylhydrolase (PAF-AH, lipoprotein associated phospholipase A2). Cardiovasc. Drugs Ther. 23, 73–83 (2009).
Zhou, M. et al. Association of the G994T and R92H genotypes of platelet-activating factor acetylhydrolase with risk of preeclampsia in Chinese women. Pregnancy Hypertens. 20, 19–26 (2020).
Gregson, J. M. et al. Genetic invalidation of Lp-PLA2 as a therapeutic target: large-scale study of five functional Lp-PLA2-lowering alleles. Eur. J. Prev. Cardiol. 24, 492–504 (2017).
Drenos, F. et al. Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum. Mol. Genet. 18, 2305–2316 (2009).
Grallert, H. et al. Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: meta-analysis of genome-wide association studies from five community-based studies. Eur. Heart J. 33, 238–251 (2012).
Liu, Y. et al. Effects of genetic variants on lipid parameters and dyslipidemia in a Chinese population. J. Lipid Res. 52, 354–360 (2011).
Tsimihodimos, V. et al. Altered distribution of platelet-activating factor- acetylhydrolase activity between LDL and HDL as a function of the severity of hypercholesterolemia. J. Lipid Res. 43, 256–263 (2002).
Tselepis, A. D., Dentan, C., Karabina, S. A., Chapman, M. J. & Ninio, E. PAF-degrading acetylhydrolase is preferentially associated with dense LDL and VHDL-1 in human plasma. Catalytic characteristics and relation to the monocyte-derived enzyme. Arterioscler. Thromb. Vasc. Biol. 15, 1764–1773 (1995).
Benitez, S. et al. Platelet-activating factor acetylhydrolase is mainly associated with electronegative low-density lipoprotein subfraction. Circulation 108, 92–96 (2003).
Pruzanski, W. & Vadas, P. Secretory synovial fluid phospholipase A2 and its role in the pathogenesis of inflammation in arthritis. J. Rheumatol. 15, 1601–1603 (1988).
Dennis, E. A., Cao, J., Hsu, Y. H., Magrioti, V. & Kokotos, G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111, 6130–6185 (2011).
Murakami, M., Taketomi, Y., Sato, H. & Yamamoto, K. Secreted phospholipase A2 revisited. J. Biochem. 150, 233–255 (2011).
Akinkuolie, A. O. et al. Group IIA secretory phospholipase A2, vascular inflammation, and incident cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 39, 1182–1190 (2019).
Breitling, L. P. et al. Type II secretory phospholipase A2 and prognosis in patients with stable coronary heart disease: Mendelian randomization study. PLoS ONE 6, e22318 (2011).
Lemaitre, R. N. et al. Circulating n-3 fatty acids and trans-fatty acids, PLA2G2A gene variation and sudden cardiac arrest. J. Nutr. Sci. 5, e12 (2016).
Andersen, V., Halekoh, U., Tjonneland, A., Vogel, U. & Kopp T. I. Intake of red and processed meat, use of non-steroid anti-inflammatory drugs, genetic variants and risk of colorectal cancer: a prospective study of the Danish “Diet, Cancer and Health” cohort. Int. J. Mol. Sci. 20, 1121 (2019).
Lands, W. E. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 231, 883–888 (1958).
Acknowledgements
The study was supported by the National Natural Science Foundation of China (Grant No. 81973053), and discipline construction funding of public health and preventive medicine from Peking University Health Science Center (BMU2020XY010).
Author information
Authors and Affiliations
Contributions
H.-J.W. and H.W. concepted and designed the study. H.W. and Y.W. wrote the draft of the manuscript. H.W., Y.W., J.Y.S., Q-Y.S., C.-X.L., and L.L. analyzed data and P.-P.Z. checked and repeated all the analyzing steps and checked all the original data. J.-Y.S., Q.-Y.S., C.-X.L., L.L., and P.-P.Z. collected all the samples and extracted the DNA from the samples. H.-J.W. improved the writing of the manuscript and all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
The informed consent was required for the present study and all the informed consent were signed by participants and their parents.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, H., Wang, Y., Song, JY. et al. Associations of genetic variants of lysophosphatidylcholine metabolic enzymes with levels of serum lipids. Pediatr Res 91, 1595–1599 (2022). https://doi.org/10.1038/s41390-021-01549-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01549-9
- Springer Nature America, Inc.
This article is cited by
-
Association of BMAL1 clock gene polymorphisms with fasting glucose in children
Pediatric Research (2023)
-
Protective Association of APOC1/rs4420638 with Risk of Obesity: A case-control Study in Portuguese Children
Biochemical Genetics (2023)
-
A simple preparation step to remove excess liquid lipids in white adipose tissue enabling improved detection of metabolites via MALDI-FTICR imaging MS
Histochemistry and Cell Biology (2022)